7OCJ
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS2(Nb14509) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, NbSOS2 (14509) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2021-04-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
4WVP
 
 | | Crystal structure of an activity-based probe HNE complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BTN-3V3-NLB-OMT-OIC-3V2, ... | | Authors: | Lechtenberg, B.C, Kasperkiewicz, P, Robinson, H.R, Drag, M, Riedl, S.J. | | Deposit date: | 2014-11-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Elastase-PK101 Structure: Mechanism of an Ultrasensitive Activity-based Probe Revealed.
Acs Chem.Biol., 10, 2015
|
|
4W97
 
 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | Authors: | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
1PPG
 
 | | The refined 2.3 angstroms crystal structure of human leukocyte elastase in a complex with a valine chloromethyl ketone inhibitor | | Descriptor: | HUMAN LEUKOCYTE ELASTASE, MEO-SUCCINYL-ALA-ALA-PRO-VAL CHLOROMETHYLKETONE, alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bode, W, Wei, A-Z. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The refined 2.3 A crystal structure of human leukocyte elastase in a complex with a valine chloromethyl ketone inhibitor.
FEBS Lett., 234, 1988
|
|
4U4R
 
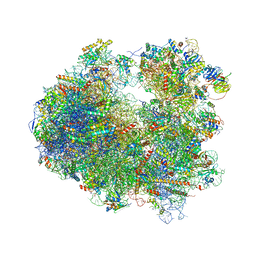 | | Crystal structure of Lactimidomycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R,5S,6E)-2-hydroxy-5-methyl-7-[(2R,3S,4E,6Z,10E)-3-methyl-12-oxooxacyclododeca-4,6,10-trien-2-yl]-4-oxooct-6-en-1-yl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
7TEA
 
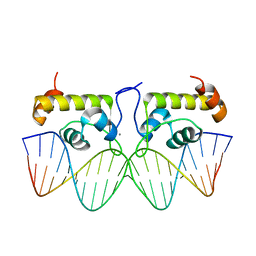 | | Crystal structure of S. aureus GlnR-DNA complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*AP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*TP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-01-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
8D8I
 
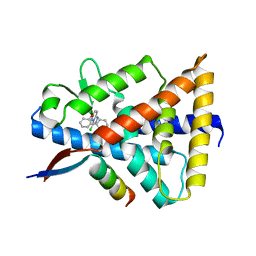 | | Crystal structure of Reverb alpha in complex with synthetic agonist | | Descriptor: | (4S)-6-[([1,1'-biphenyl]-2-yl)oxy]-3-chloro[1,2,4]triazolo[4,3-b]pyridazine, Nuclear receptor corepressor 1, Nuclear receptor subfamily 1 group D member 1 | | Authors: | Ronin, C, Ciesielski, F, Hegazy, L, Burris, P.T. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis of synthetic agonist activation of the nuclear receptor REV-ERB.
Nat Commun, 13, 2022
|
|
4WWC
 
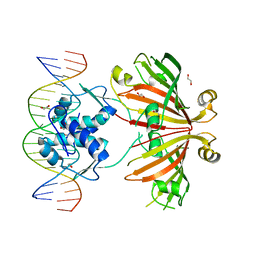 | | Crystal structure of full length YvoA in complex with palindromic operator DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*AP*GP*TP*GP*GP*TP*CP*TP*AP*GP*AP*CP*CP*AP*CP*TP*GP*G)-3'), HTH-type transcriptional repressor YvoA | | Authors: | Grau, F.C, Fillenberg, S.B, Muller, Y.A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural insight into operator dre-sites recognition and effector binding in the GntR/HutC transcription regulator NagR.
Nucleic Acids Res., 43, 2015
|
|
3JSP
 
 | |
1DGD
 
 | |
1DGE
 
 | |
8R7Y
 
 | | Deoxyribonucleoside regulator DeoR in complex with the DNA operator | | Descriptor: | Deoxyribonucleoside regulator, OL18 DNA operator, strand 1, ... | | Authors: | Pachl, P, Soltysova, M, Rezacova, P. | | Deposit date: | 2023-11-27 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural characterization of two prototypical repressors of SorC family reveals tetrameric assemblies on DNA and mechanism of function.
Nucleic Acids Res., 52, 2024
|
|
5KWQ
 
 | | Two Tandem RRM Domains of FBP-Interacting Repressor (FIR), also Known as PUF60 | | Descriptor: | Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Yang, Y, Zhou, H, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-18 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
7ET5
 
 | | Crystal structure of Arabidopsis TEM1 AP2 domain | | Descriptor: | AP2/ERF and B3 domain-containing transcription repressor TEM1, SULFATE ION | | Authors: | Hu, H, Du, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ET6
 
 | | Crystal structure of Arabidopsis TEM1 B3-DNA complex | | Descriptor: | AP2/ERF and B3 domain-containing transcription repressor TEM1, FT-RY14-F, FT-RY14-R | | Authors: | Hu, H, Du, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ET4
 
 | | Crystal structure of Arabidopsis TEM1 AP2 domain | | Descriptor: | AP2/ERF and B3 domain-containing transcription repressor TEM1, DNA (12-mer) | | Authors: | Hu, H, Du, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1ZAY
 
 | | PURINE REPRESSOR-HYPOXANTHINE-MODIFIED-PURF-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*TP*(1AP)P*TP*TP*TP*TP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Zheleznova, E.E, Brennan, R.G. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Roles of Exocyclic Groups in the Central Base-Pair Step in Modulating the Affinity of PurR for its Operator
To be Published
|
|
7CNA
 
 | | Crystal structure of Spindlin1/C11orf84 complex bound to histone H3K4me3K9me3 peptide | | Descriptor: | ALA-ARG-THR-M3L-GLN-THR-ALA-ARG-M3L-SER-GLY, ALA-ARG-THR-M3L-GLN-THR-ALA-ARG-M3L-SER-THR, BENZAMIDINE, ... | | Authors: | Qian, C.M. | | Deposit date: | 2020-07-30 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural mechanism of bivalent histone H3K4me3K9me3 recognition by the Spindlin1/C11orf84 complex in rRNA transcription activation.
Nat Commun, 12, 2021
|
|
7ZRA
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS1(Nb14497) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody NbSOS1 (Nb14497) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2022-05-04 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
6ZUI
 
 | | Crystal structure of the Cys-Ser mutant of the cpYFP-based biosensor for hypochlorous acid | | Descriptor: | HTH-type transcriptional repressor NemR,Green fluorescent protein,Green fluorescent protein,HTH-type transcriptional repressor NemR | | Authors: | Tossounian, M.A, Van Molle, I, Messens, J. | | Deposit date: | 2020-07-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.200082 Å) | | Cite: | Hypocrates is a genetically encoded fluorescent biosensor for (pseudo)hypohalous acids and their derivatives.
Nat Commun, 13, 2022
|
|
7RMW
 
 | | Crystal structure of B. subtilis PurR bound to ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Pur operon repressor | | Authors: | Schumacher, M.A. | | Deposit date: | 2021-07-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The nucleotide messenger (p)ppGpp is an anti-inducer of the purine synthesis transcription regulator PurR in Bacillus.
Nucleic Acids Res., 50, 2022
|
|
4WLS
 
 | | Crystal structure of the metal-free (repressor) form of E. Coli CUER, a copper efflux regulator, bound to COPA promoter DNA | | Descriptor: | COPA PROMOTER DNA NON-TEMPLATE STRAND, COPA PROMOTER DNA NON-TEMPLATE STRAND (ALTERNATE CONFORMATION), COPA PROMOTER DNA TEMPLATE STRAND, ... | | Authors: | Philips, S.J, Canalizo-Hernandez, M, Mondragon, A, O'Halloran, T.V. | | Deposit date: | 2014-10-08 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Allosteric transcriptional regulation via changes in the overall topology of the core promoter.
Science, 349, 2015
|
|
6FAS
 
 | | Crystal structure of VAL1 B3 domain in complex with cognate DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*GP*CP*CP*AP*TP*GP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*TP*GP*CP*AP*TP*GP*GP*CP*T)-3') | | Authors: | Sasnauskas, G. | | Deposit date: | 2017-12-17 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of DNA target recognition by the B3 domain of Arabidopsis epigenome reader VAL1.
Nucleic Acids Res., 46, 2018
|
|
7ZPN
 
 | | Crystal Structure of IscR from Dinoroseobacter shibae | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator, SULFATE ION | | Authors: | Lukat, P, Ploetzky, L, Blankenfeldt, W, Jahn, D, Haertig, E. | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dinoroseobacter shibae IscR homolog acts as a repressor for iron acquisition genes
To Be Published
|
|
5YEL
 
 | | Crystal structure of CTCF ZFs6-11-gb7CSE | | Descriptor: | DNA (26-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-18 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
