8OOY
 
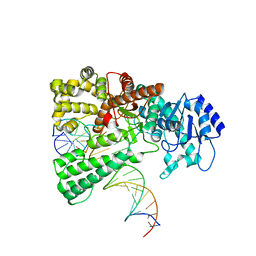 | | Pol I bound to extended and displaced DNA section - open conformation | | Descriptor: | DNA polymerase I, Displacing Primer, Extending Primer, ... | | Authors: | Botto, M, Borsellini, A, Lamers, M.H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8TLT
 
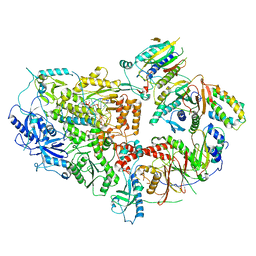 | | Cryo-EM structure of Rev1(deltaN)-Polzeta-DNA-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*TP*AP*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*AP*AP*T)-3'), ... | | Authors: | Malik, R, Aggarwal, A.K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the Rev1-Pol zeta holocomplex reveals the mechanism of their cooperativity in translesion DNA synthesis.
Nat.Struct.Mol.Biol., 2024
|
|
8TLQ
 
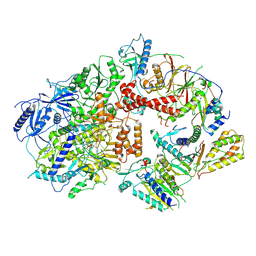 | | Cryo-EM structure of the Rev1-Polzeta-DNA-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Malik, R, Aggarwal, A.K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structure of the Rev1-Pol zeta holocomplex reveals the mechanism of their cooperativity in translesion DNA synthesis.
Nat.Struct.Mol.Biol., 2024
|
|
4HRE
 
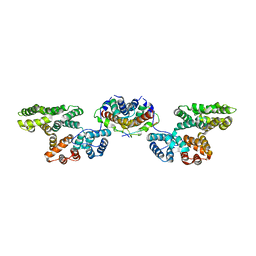 | | Crystal Structure of p11/Annexin A2 Heterotetramer in Complex with SMARCA3 Peptide | | Descriptor: | Annexin A2, Helicase-like transcription factor, Protein S100-A10 | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2012-10-27 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7852 Å) | | Cite: | SMARCA3, a Chromatin-Remodeling Factor, Is Required for p11-Dependent Antidepressant Action.
Cell(Cambridge,Mass.), 152, 2013
|
|
8USR
 
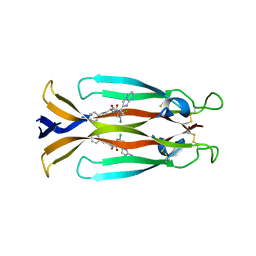 | | IL17A homodimer complexed to Compound 23 | | Descriptor: | Interleukin-17A, ~{N}-[(2~{S})-1-[[(1~{S})-1-(8~{a}~{H}-imidazo[1,2-a]pyrimidin-2-yl)ethyl]amino]-1-oxidanylidene-4-phenyl-butan-2-yl]-4,5-bis(chloranyl)-1~{H}-pyrrole-2-carboxamide | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 67, 2024
|
|
8USS
 
 | | IL17A complexed to Compound 7 | | Descriptor: | 4,5-dichloro-N-[(1S)-1-cyclohexyl-2-{[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]amino}-2-oxoethyl]-1H-pyrrole-2-carboxamide, CHLORIDE ION, Interleukin-17A | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 67, 2024
|
|
5V7K
 
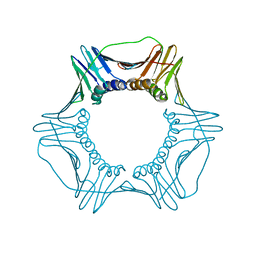 | |
1N54
 
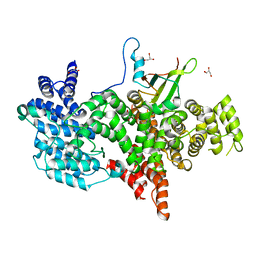 | | Cap Binding Complex m7GpppG free | | Descriptor: | 20 kDa nuclear cap binding protein, 80 kDa nuclear cap binding protein, GLYCEROL | | Authors: | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | Deposit date: | 2002-11-04 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
8YBX
 
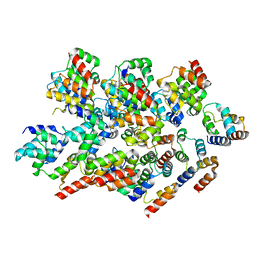 | |
8YD8
 
 | |
8YD7
 
 | |
5M2N
 
 | |
8QFC
 
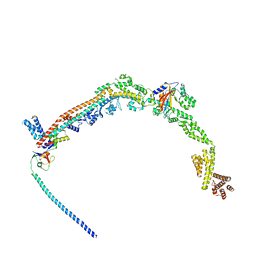 | | UFL1 E3 ligase bound 60S ribosome | | Descriptor: | 60S ribosomal protein L10a, CDK5 regulatory subunit-associated protein 3, DDRGK domain-containing protein 1, ... | | Authors: | Makhlouf, L, Zeqiraj, E, Kulathu, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
2DR5
 
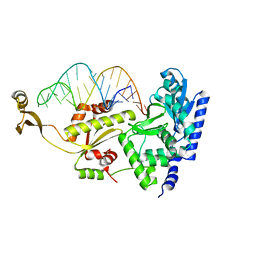 | | Complex structure of CCA adding enzyme with mini-helix lacking CCA | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (32-MER) | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DVI
 
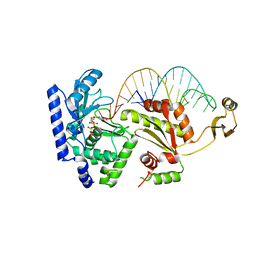 | | Complex structure of CCA-adding enzyme, mini-DCC and CTP | | Descriptor: | CCA-adding enzyme, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-07-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
5LLI
 
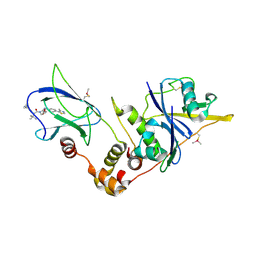 | | pVHL:EloB:EloC in complex with VH298 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-cyanocyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent and selective chemical probe of hypoxic signalling downstream of HIF-alpha hydroxylation via VHL inhibition.
Nat Commun, 7, 2016
|
|
7YI1
 
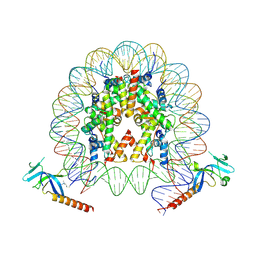 | | Cryo-EM structure of Eaf3 CHD bound to H3K36me3 nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
5OFN
 
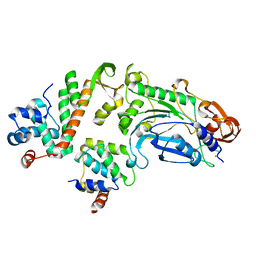 | |
5OF3
 
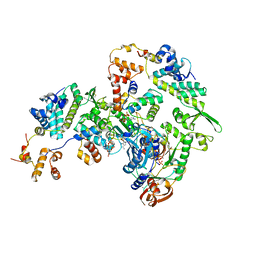 | |
8B9C
 
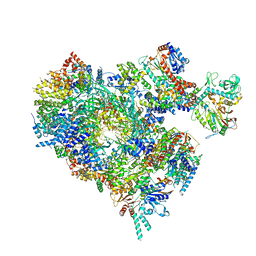 | | S. cerevisiae pol alpha - replisome complex | | Descriptor: | Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA polymerase alpha catalytic subunit A, ... | | Authors: | Jones, M.L, Yeeles, J.T.P. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | How Pol alpha-primase is targeted to replisomes to prime eukaryotic DNA replication.
Mol.Cell, 83, 2023
|
|
8B9B
 
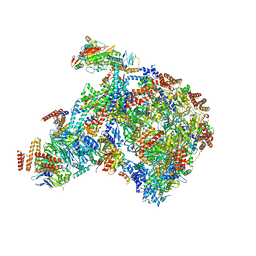 | |
8E9A
 
 | | Crystal structure of AsfvPolX in complex with 10-23 DNAzyme and Mg | | Descriptor: | DNA/RNA (52-MER), MAGNESIUM ION, Repair DNA polymerase X, ... | | Authors: | Cramer, E.R, Robart, A.R, Kaya, A.I. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of a 10-23 deoxyribozyme exhibiting a homodimer conformation.
Commun Chem, 6, 2023
|
|
8B9A
 
 | |
5BSA
 
 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.611 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
7F60
 
 | |
