4NRA
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with compound-6 E11322 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-chloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl)ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Ferguson, F.M, Filippakopoulos, P, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain.
J.Med.Chem., 56, 2013
|
|
4G8J
 
 | | X-ray Structure of Uridine Phosphorylease from Vibrio cholerae Complexed with Thymidine at 2.12 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Lashkov, A.A, Gabdoulkhakov, A.G, Prokofev, I.I, Sotnichenko, S.E, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-07-23 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | X-ray structure of uridine phosphorylease from Vibrio cholerae complexed with thymidine
To be Published
|
|
3EXU
 
 | | A glycoside hydrolase family 11 xylanase with an extended thumb region | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase, GLYCEROL | | Authors: | Vandermarliere, E, Pollet, A, Strelkov, S.V, Delcour, J.A, Courtin, C.M. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic and activity-based evidence for thumb flexibility and its relevance in glycoside hydrolase family 11 xylanases
Proteins, 77, 2009
|
|
1HG5
 
 | | CALM-N N-terminal domain of clathrin assembly lymphoid myeloid leukaemia protein, inositol(1,2,3,4,5,6)P6 complex | | Descriptor: | CLATHRIN ASSEMBLY PROTEIN SHORT FORM, INOSITOL HEXAKISPHOSPHATE | | Authors: | Ford, M.G.J, Evans, P.R, McMahon, H.T. | | Deposit date: | 2000-12-12 | | Release date: | 2001-02-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Simultaneous Binding of Ptdins(4,5)P2 and Clathrin by Ap180 in the Nucleation of Clathrin Lattices on Membranes
Science, 291, 2001
|
|
1OUK
 
 | | The structure of p38 alpha in complex with a pyridinylimidazole inhibitor | | Descriptor: | 4-[5-[2-(1-PHENYL-ETHYLAMINO)-PYRIMIDIN-4-YL]-1-METHYL-4-(3-TRIFLUOROMETHYLPHENYL)-1H-IMIDAZOL-2-YL]-PIPERIDINE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
4K5Z
 
 | | Crystal Structure of Human Chymase in Complex with Fragment Inhibitor 6-chloro-2,3-dihydro-1H-isoindol-1-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-2,3-dihydro-1H-isoindol-1-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
3I80
 
 | | Protein Tyrosine Phosphatase 1B - Transition state analog for the second catalytic step | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, ... | | Authors: | Brandao, T.A.S, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2009-07-09 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into the reaction of protein-tyrosine phosphatase 1B: crystal structures for transition state analogs of both catalytic steps.
J.Biol.Chem., 285, 2010
|
|
1P9P
 
 | | The Crystal Structure of a M1G37 tRNA Methyltransferase, TrmD | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Watts, J.M, Zalacain, M, Van Thiel, A, Vitaszka, P.R, Redlak, M, Andraos-Selim, C, Rastinejad, F, Holmes, W.M. | | Deposit date: | 2003-05-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into Catalysis by a Knotted TrmD tRNA Methyltransferase.
J.Mol.Biol., 333, 2003
|
|
3CKE
 
 | | Crystal structure of aristolochene synthase in complex with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Yu, F, Miller, D.J, Faraldos, J.A, Zhao, Y, Coates, R.M, Allemann, R.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis.
J.Biol.Chem., 283, 2008
|
|
2MWM
 
 | |
4GCC
 
 | |
4ZWC
 
 | | Crystal structure of maltose-bound human GLUT3 in the outward-open conformation at 2.6 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
3LZM
 
 | |
4ZZH
 
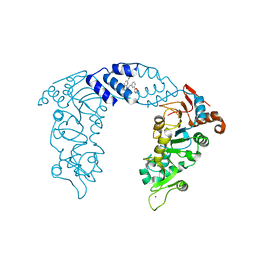 | | SIRT1/Activator Complex | | Descriptor: | (4S)-N-[3-(1,3-oxazol-5-yl)phenyl]-7-[3-(trifluoromethyl)phenyl]-3,4-dihydro-1,4-methanopyrido[2,3-b][1,4]diazepine-5(2H)-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ZINC ION | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
4KDP
 
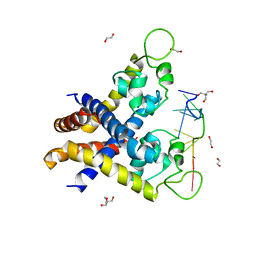 | | TcaR-ssDNA complex crystal structure reveals the novel ssDNA binding mechanism of the MarR family proteins | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(*CP*GP*CP*AP*GP*CP*GP*CP*GP*CP*AP*GP*CP*CP*CP*TP*A)-3'), ... | | Authors: | Chang, Y.M, Chen, C.K.-M, Wang, A.H.-J. | | Deposit date: | 2013-04-25 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | TcaR-ssDNA complex crystal structure reveals new DNA binding mechanism of the MarR family proteins.
Nucleic Acids Res., 42, 2014
|
|
4FTI
 
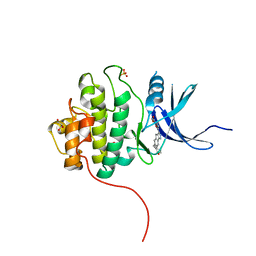 | | Crystal Structure of the CHK1 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FTR
 
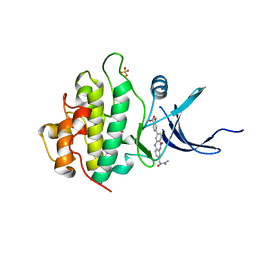 | | Crystal Structure of the CHK1 | | Descriptor: | 2-[3-(3-methoxy-4-nitrophenyl)-11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-8-yl]-N,N-dimethylacetamide, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
3IGB
 
 | | Bace-1 with Compound 3 | | Descriptor: | 8,8-diphenyl-2,3,4,8-tetrahydroimidazo[1,5-a]pyrimidin-6-amine, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Aminoimidazoles as potent and selective human beta-secretase (BACE1) inhibitors
J.Med.Chem., 52, 2009
|
|
3IE9
 
 | | Structure of oxidized M98L mutant of amicyanin | | Descriptor: | ACETATE ION, Amicyanin, CHLORIDE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining the role of the axial ligand of the type 1 copper site in amicyanin by replacement of methionine with leucine.
Biochemistry, 48, 2009
|
|
1OYF
 
 | | Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol | | Descriptor: | 6-METHYLHEPTAN-1-OL, ACETIC ACID, Phospholipase A2, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-04-04 | | Release date: | 2003-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol
To be Published
|
|
1LL6
 
 | |
4ZZX
 
 | | Structure of PARP2 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-(3-methoxypropyl)-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, POLY [ADP-RIBOSE] POLYMERASE 2 | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
5QI2
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000110a | | Descriptor: | 1-methyl-3-[3-(2-methylpyrimidin-4-yl)phenyl]urea, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4JVB
 
 | | Crystal structure of PDE6D in complex with the inhibitor rac-2 | | Descriptor: | 1-benzyl-2-(4-{[(2R)-2-(2-phenyl-1H-benzimidazol-1-yl)pent-4-en-1-yl]oxy}phenyl)-1H-benzimidazole, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Gunther, Z, Papke, B, Ismail, S, Vartak, N, Chandra, A, Hoffmann, M, Hahn, S, Triola, G, Wittinghofer, A, Bastiaens, P, Waldmann, H. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small molecule inhibition of the KRAS PDEd interaction impairs oncogenic KRAS signalling
Nature, 497, 2013
|
|
2XP2
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with Crizotinib (PF-02341066) | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, TYROSINE-PROTEIN KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Timofeevski, S, Marrone, T, Cui, J.J. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure Based Drug Design of Crizotinib (Pf-02341066), a Potent and Selective Dual Inhibitor of Mesenchymal-Epithelial Transition Factor (C-met) Kinase and Anaplastic Lymphoma Kinase (Alk).
J.Med.Chem, 54, 2011
|
|
