8WNI
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
5N3J
 
 | | cAMP-dependent Protein Kinase A from Cricetulus griseus in complex with fragment like molecule 4-Nitrobenzoic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-NITROBENZOIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 2020
|
|
5N3Q
 
 | | cAMP-dependent Protein Kinase A from Cricetulus griseus in complex with fragment like molecule 3-Aminobenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-aminobenzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 2020
|
|
5N3H
 
 | |
5N3S
 
 | | cAMP-dependent Protein Kinase A from Cricetulus griseus in complex with fragment like molecule 4-Hydroxybenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-HYDROXYBENZAMIDE, DIMETHYL SULFOXIDE, ... | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 2020
|
|
2P9V
 
 | | Structure of AmpC beta-lactamase with cross-linked active site after exposure to small molecule inhibitor | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Babaoglu, K, Wyrembak, P.N, Pelto, R.B, Shoichet, B.K, Pratt, R.F. | | Deposit date: | 2007-03-26 | | Release date: | 2007-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | O-Aryloxycarbonyl Hydroxamates: New beta-Lactamase Inhibitors That Cross-Link the Active Site.
J.Am.Chem.Soc., 129, 2007
|
|
4E3J
 
 | |
1M5Z
 
 | | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area | | Descriptor: | AMPA receptor interacting protein | | Authors: | Feng, W, Fan, J, Jiang, M, Shi, Y, Zhang, M. | | Deposit date: | 2002-07-11 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area
J.Biol.Chem., 277, 2002
|
|
7NP3
 
 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
2P98
 
 | | E. coli methionine aminopeptidase monometalated with inhibitor YE7 | | Descriptor: | IMIDAZO[2,1-A]ISOQUINOLINE-2-CARBOHYDRAZIDE, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q. | | Deposit date: | 2007-03-24 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Monometalated Methionine Aminopeptidase: Inhibitor Discovery and Crystallographic Analysis.
J.Med.Chem., 50, 2007
|
|
4X68
 
 | | Crystal Structure of OP0595 complexed with AmpC | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, NICKEL (II) ION | | Authors: | Yamada, M, Watanabe, T. | | Deposit date: | 2014-12-07 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
5C76
 
 | |
6EJI
 
 | | Structure of a glycosyltransferase | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
6EJK
 
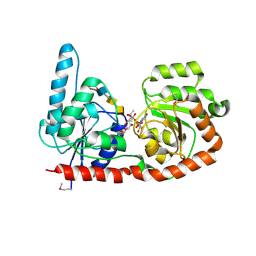 | | Structure of a glycosyltransferase | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, NerylNeryl pyrophosphate, Uridine-Diphosphate-Methylene-N-acetyl-galactosamine, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
7SAY
 
 | |
4W9M
 
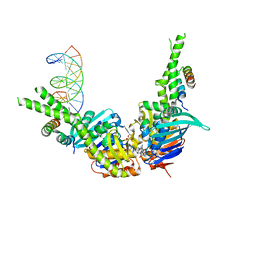 | | AMPPNP bound Rad50 in complex with dsDNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*GP*TP*CP*AP*CP*CP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*GP*GP*TP*GP*AP*CP*CP*GP*AP*CP*C)-3'), Exonuclease, ... | | Authors: | Rojowska, A, Lammens, K. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Rad50 DNA double-strand break repair protein in complex with DNA.
Embo J., 33, 2014
|
|
5GXG
 
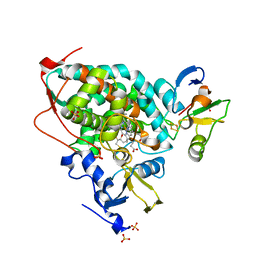 | | High-resolution crystal structure of the electron transfer complex of cytochrome p450cam with putidaredoxin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kikui, Y, Hiruma, Y, Ubbink, M, Nojiri, M. | | Deposit date: | 2016-09-17 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of productive and futile encounters in an electron transfer protein complex
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2K6O
 
 | | Human LL-37 Structure | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Wang, G. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles
J.Biol.Chem., 283, 2008
|
|
2LQD
 
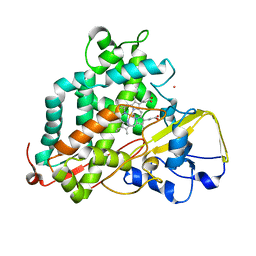 | | Reduced and CO-bound cytochrome P450cam (CYP101A1) | | Descriptor: | CARBON MONOXIDE, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Pochapsky, T.C, Pochapsky, S.S, Asciutto, E, Madura, J, Young, M.J. | | Deposit date: | 2012-03-02 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structural Ensembles of Substrate-Free Cytochrome P450(cam).
Biochemistry, 51, 2012
|
|
5OWV
 
 | |
6VXJ
 
 | | Structure of ABCG2 bound to SN38 | | Descriptor: | 7-ethyl-10-hydroxycamptothecin, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL | | Authors: | Orlando, B.J, Liao, M. | | Deposit date: | 2020-02-22 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
8CY8
 
 | |
2LMF
 
 | | Solution structure of human LL-23 bound to membrane-mimetic micelles | | Descriptor: | Antibacterial protein LL-37 | | Authors: | Wang, G. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Dynamics, and Antimicrobial and Immune Modulatory Activities of Human LL-23 and Its Single-Residue Variants Mutated on the Basis of Homologous Primate Cathelicidins.
Biochemistry, 51, 2012
|
|
2J9A
 
 | | blLAP in Complex with Microginin FR1 | | Descriptor: | (2S,3R)-3-AMINO-2-HYDROXYDECANOIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Kraft, M, Schleberger, C, Weckesser, J, Schulz, G.E. | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Binding Structure of the Leucine Aminopeptidase Inhibitor Microginin Fr1.
FEBS Lett., 580, 2006
|
|
5NMN
 
 | |
