1ESZ
 
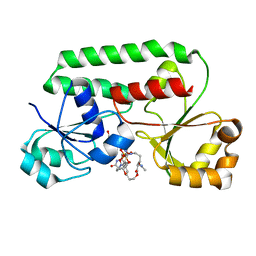 | | STRUCTURE OF THE PERIPLASMIC FERRIC SIDEROPHORE BINDING PROTEIN FHUD COMPLEXED WITH COPROGEN | | Descriptor: | COPROGEN, FERRICHROME-BINDING PERIPLASMIC PROTEIN | | Authors: | Clarke, T.E, Braun, V, Winkelmann, G, Tari, L.W, Vogel, H.J. | | Deposit date: | 2000-04-11 | | Release date: | 2002-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin.
J.Biol.Chem., 277, 2002
|
|
2OXL
 
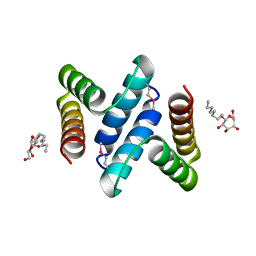 | | Structure and Function of the E. coli Protein YmgB: a Protein Critical for Biofilm Formation and Acid Resistance | | Descriptor: | Hypothetical protein ymgB, octyl beta-D-glucopyranoside | | Authors: | Page, R, Peti, W, Woods, T.K, Palermino, J.M, Doshi, O. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the Escherichia coli Protein YmgB: A Protein Critical for Biofilm Formation and Acid-resistance.
J.Mol.Biol., 373, 2007
|
|
1ET7
 
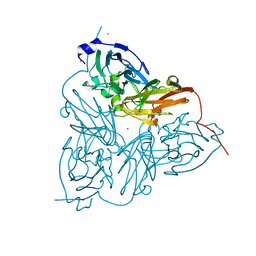 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASP MUTANT FROM ALCALIGENES FAECALIS S-6 | | Descriptor: | CADMIUM ION, COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
5LID
 
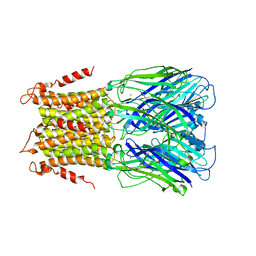 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromopromazine | | Descriptor: | Cys-loop ligand-gated ion channel, bromopromazine | | Authors: | Nys, M, Wijckmans, E, Farinha, A, Brams, M, Spurny, R, Ulens, C. | | Deposit date: | 2016-07-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1EJ3
 
 | | CRYSTAL STRUCTURE OF AEQUORIN | | Descriptor: | AEQUORIN, C2-HYDROPEROXY-COELENTERAZINE | | Authors: | Head, J.F, Inouye, S, Teranishi, K, Shimomura, O. | | Deposit date: | 2000-02-29 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the photoprotein aequorin at 2.3 A resolution.
Nature, 405, 2000
|
|
1EJF
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CO-CHAPERONE P23 | | Descriptor: | Prostaglandin E synthase 3, SULFATE ION | | Authors: | Weaver, A.J, Sullivan, W.P, Felts, S.J, Owen, B.A.L, Toft, D.O. | | Deposit date: | 2000-03-02 | | Release date: | 2000-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone.
J.Biol.Chem., 275, 2000
|
|
5LKM
 
 | | RadA bound to dTDP | | Descriptor: | DNA repair protein RadA, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Rapisarda, C, Remaut, H, Fornzes, R. | | Deposit date: | 2016-07-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension.
Nat Commun, 8, 2017
|
|
1EJN
 
 | | UROKINASE PLASMINOGEN ACTIVATOR B-CHAIN INHIBITOR COMPLEX | | Descriptor: | N-(1-ADAMANTYL)-N'-(4-GUANIDINOBENZYL)UREA, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Sperl, S, Jacob, U, Arroyo de Prada, N, Stuerzebecher, J, Wilhelm, O.G, Bode, W, Magdolen, V, Huber, R, Moroder, L. | | Deposit date: | 2000-04-22 | | Release date: | 2000-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | (4-aminomethyl)phenylguanidine derivatives as nonpeptidic highly selective inhibitors of human urokinase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EJR
 
 | | CRYSTAL STRUCTURE OF THE D221A VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | NICKEL (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Pearson, M.A, Park, I.S, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-03-04 | | Release date: | 2000-09-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of urease active site variants.
Biochemistry, 39, 2000
|
|
5LL1
 
 | |
1EU6
 
 | |
1EUJ
 
 | | A NOVEL ANTI-TUMOR CYTOKINE CONTAINS A RNA-BINDING MOTIF PRESENT IN AMINOACYL-TRNA SYNTHETASES | | Descriptor: | ENDOTHELIAL MONOCYTE ACTIVATING POLYPEPTIDE 2 | | Authors: | Kim, Y, Shin, J, Li, R, Cheong, C, Kim, S. | | Deposit date: | 2000-04-17 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel anti-tumor cytokine contains an RNA binding motif present in aminoacyl-tRNA synthetases.
J.Biol.Chem., 275, 2000
|
|
5LLM
 
 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with L-ASP and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-27 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
1EK5
 
 | | STRUCTURE OF HUMAN UDP-GALACTOSE 4-EPIMERASE IN COMPLEX WITH NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GALACTOSE 4-EPIMERASE | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase.
Biochemistry, 39, 2000
|
|
5LLT
 
 | | Plasmodium falciparum nicotinic acid mononucleotide adenylyltransferase complexed with NaAD | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINIC ACID ADENINE DINUCLEOTIDE, ... | | Authors: | Bathke, J, Fritz-Wolf, K, Brandstaedter, C, Rahlfs, S, Becker, K. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Characterization of Plasmodium falciparum Nicotinic Acid Mononucleotide Adenylyltransferase.
J. Mol. Biol., 428, 2016
|
|
1EKK
 
 | | CRYSTAL STRUCTURE OF HYDROXYETHYLTHIAZOLE KINASE IN THE R3 FORM WITH HYDROXYETHYLTHIAZOLE | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, HYDROXYETHYLTHIAZOLE KINASE, SULFUR DIOXIDE | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-03-09 | | Release date: | 2000-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
5LM1
 
 | | Crystal Structure of HD-PTP phosphatase in complex with UBAP1 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 23, UBAP-1 | | Authors: | Levy, C. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for Selective Interaction between the ESCRT Regulator HD-PTP and UBAP1.
Structure, 24, 2016
|
|
1EKZ
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE THIRD DSRBD FROM DROSOPHILA STAUFEN AND A RNA HAIRPIN | | Descriptor: | MATERNAL EFFECT PROTEIN (STAUFEN), STAUFEN DOUBLE-STRANDED RNA BINDING DOMAIN | | Authors: | Ramos, A, Grunert, S, Bycroft, M, St Johnston, D, Varani, G. | | Deposit date: | 2000-03-11 | | Release date: | 2000-08-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | RNA recognition by a Staufen double-stranded RNA-binding domain.
EMBO J., 19, 2000
|
|
9GEW
 
 | | Crystal structure of CREBBP bromodomain in complex with (R,E)-6-(5-(6-methoxy-2,3-dihydro-4H-benzo[b][1,4]oxazin-4-yl)pent-1-en-1-yl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one | | Descriptor: | (4~{R})-6-[(~{E})-5-(6-methoxy-2,3-dihydro-1,4-benzoxazin-4-yl)pent-1-enyl]-4-methyl-1,3,4,5-tetrahydro-1,5-benzodiazepin-2-one, (R,R)-2,3-BUTANEDIOL, CREBBP | | Authors: | Amann, M, Boyd, A, Einsle, O, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of CREBBP bromodomain in complex with (R,E)-6-(5-(6-methoxy-2,3-dihydro-4H-benzo[b][1,4]oxazin-4-yl)pent-1-en-1-yl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one
To Be Published
|
|
5LMV
 
 | | Structure of bacterial 30S-IF1-IF2-IF3-mRNA-tRNA translation pre-initiation complex(state-III) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
9GFA
 
 | | Crystal structure of 14-3-3 sigma in complex with Tau pS214 peptide and covalent stabilizer LD33 | | Descriptor: | 14-3-3 protein sigma, 3-(3-bromanyl-4-methyl-phenyl)-2-pyridin-3-yl-1,2-dihydrobenzimidazole, Microtubule-associated protein tau | | Authors: | van den Oetelaar, M.C.M, Engelen, S.F.H, Ottmann, C, Brunsveld, L. | | Deposit date: | 2024-08-08 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Site-selective stabilization of the 14-3-3/tau protein-protein interaction
To Be Published
|
|
1EP1
 
 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
1EPO
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH CP-81,282 (MOR PHE NLE CHF NME) | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide | | Authors: | Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct observation by X-ray analysis of the tetrahedral intermediate of aspartic proteinases.
Protein Sci., 1, 1992
|
|
5LZT
 
 | | Structure of the mammalian ribosomal termination complex with eRF1 and eRF3. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
1E9Z
 
 | |
