5CD8
 
 | | Crystal structure of the NTD of Drosophila Oskar protein | | Descriptor: | GLYCEROL, Maternal effect protein oskar | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5N90
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with TTGTGGTGT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*TP*GP*TP*GP*GP*TP*GP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5CM8
 
 | |
5CM9
 
 | |
5CD9
 
 | | Crystal structure of the CTD of Drosophila Oskar protein | | Descriptor: | Maternal effect protein oskar, SULFATE ION | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A49
 
 | |
5N8R
 
 | | Crystal Structure of Drosophilia DHX36 helicase in complex with GAGCACTGC | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*GP*AP*GP*CP*AP*CP*TP*GP*C)-3') | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N9F
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with ssDNA CpG_A | | Descriptor: | CG9323, isoform A, CHLORIDE ION, ... | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.969 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
5CQQ
 
 | | Crystal structure of the Drosophila Zeste DNA binding domain in complex with DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*CP*GP*AP*GP*TP*GP*GP*AP*AP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*TP*TP*TP*CP*CP*AP*CP*TP*CP*GP*TP*TP*TP*TP*T)-3'), Regulatory protein zeste | | Authors: | Gao, G.N, Wang, M, Yang, N, Huang, Y, Xu, R.M. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Zeste-DNA Complex Reveals a New Modality of DNA Recognition by Homeodomain-Like Proteins
J.Mol.Biol., 427, 2015
|
|
3L1C
 
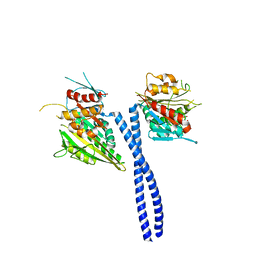 | | Kinesin-14 Protein Ncd, T436S Mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein claret segregational | | Authors: | Kull, F.J. | | Deposit date: | 2009-12-11 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A kinesin motor in a force-producing conformation.
Bmc Struct.Biol., 10, 2010
|
|
5N9A
 
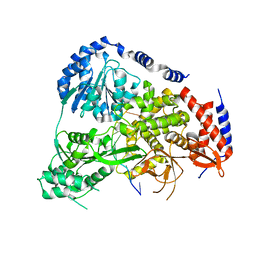 | | Crystal Structure of Drosophila DHX36 helicase in complex with GTTAGGGTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*GP*TP*TP*AP*GP*GP*GP*TP*T)-3') | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.036 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5CVE
 
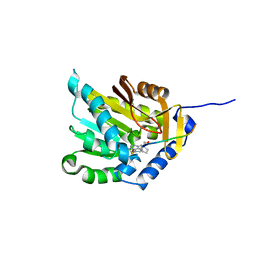 | |
3LTG
 
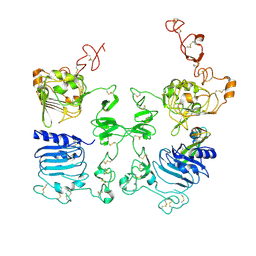 | |
5DT6
 
 | |
5AAN
 
 | | Crystal structure of Drosophila NCS-1 bound to penothiazine FD44 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CG5907-PA, ... | | Authors: | Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2015-07-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NT7
 
 | |
3LNQ
 
 | | Structure of Aristaless homeodomain in complex with DNA | | Descriptor: | 5'-D(*CP*CP*CP*TP*AP*AP*TP*TP*AP*AP*AP*CP*CP*C)-3', 5'-D(*GP*GP*GP*TP*TP*TP*AP*AP*TP*TP*AP*GP*GP*G)-3', ACETATE ION, ... | | Authors: | Takamura, Y, Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
3L9I
 
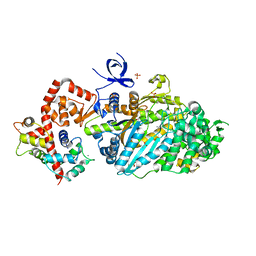 | | Myosin VI nucleotide-free (mdinsert2) L310G mutant crystal structure | | Descriptor: | ACETATE ION, CALCIUM ION, Calmodulin, ... | | Authors: | Pylypenko, O, Song, L, Sweeney, L.H, Houdusse, A. | | Deposit date: | 2010-01-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of insert i of myosin VI in modulating nucleotide affinity
To be Published
|
|
3L9K
 
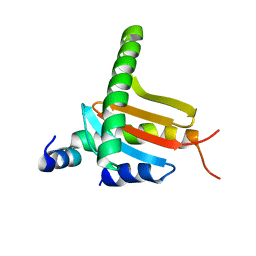 | |
3I2W
 
 | |
5E0M
 
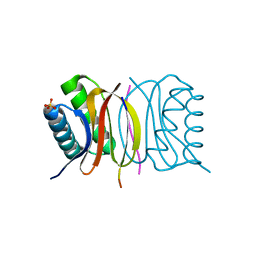 | | LC8 - Chica (468-476) Complex | | Descriptor: | Dynein light chain 1, cytoplasmic, Protein Chica peptide, ... | | Authors: | Clark, S.A, Barbar, E.B, Karplus, P.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Anchored Flexibility Model in LC8 Motif Recognition: Insights from the Chica Complex.
Biochemistry, 55, 2016
|
|
3L7H
 
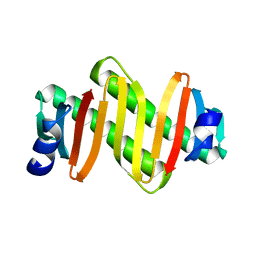 | |
4M63
 
 | | Crystal Structure of a Filament-Like Actin Trimer Bound to the Bacterial Effector VopL | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Zahm, J.A, Rosen, M.K. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | The Bacterial Effector VopL Organizes Actin into Filament-like Structures.
Cell(Cambridge,Mass.), 155, 2013
|
|
8FIA
 
 | | The structure of fly Teneurin self assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bandekar, S.J, Li, J, Arac, D. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-03 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of fly Teneurin-m reveals an asymmetric self-assembly that allows expansion into zippers.
Embo Rep., 24, 2023
|
|
4PXU
 
 | | Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bipolar kinesin KRP-130 | | Authors: | Scholey, J.E, Nithianantham, S, Scholey, J.M, Al-Bassam, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers.
Elife, 3, 2014
|
|
