7C6Z
 
 | | Crystal structure of beta-glycosides-binding protein (W67A) of ABC transporter in an open state | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C6N
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellotetraose (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFITE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C68
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to cellotetraose | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C6R
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellopentaose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
6UH9
 
 | | Crystal structure of DAD2 D166A mutant | | Descriptor: | Decreased Apical Dominance 2, TETRAETHYLENE GLYCOL | | Authors: | Sharma, P, Hamiaux, C, Snowden, K.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners.
J.Biol.Chem., 295, 2020
|
|
3OV1
 
 | |
7C69
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to sophorose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Sugar ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C6V
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to laminaritriose (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
3OPU
 
 | |
6J0O
 
 | | Crystal structure of CERT START domain in complex with compound SC1 | | Descriptor: | 2-[4-[2-fluoranyl-5-[3-(6-methylpyridin-2-yl)-1~{H}-pyrazol-4-yl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT, UNKNOWN ATOM OR ION | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-12-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
3PZE
 
 | | JNK1 in complex with inhibitor | | Descriptor: | 3-(carbamoylamino)-5-phenylthiophene-2-carboxamide, Mitogen-activated protein kinase 8, SULFATE ION | | Authors: | Xue, Y. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(piperidin-3-yl)-3-ureidothiophene-2-carboxamide (AZD7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
7VQM
 
 | | GH2 beta-galacturonate AqGalA in complex with galacturonide | | Descriptor: | CHLORIDE ION, GH2 beta-galacturonate AqGalA, beta-D-galactopyranuronic acid | | Authors: | Yang, J. | | Deposit date: | 2021-10-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Basis of a Marine Bacterial Glycoside Hydrolase Family 2 beta-Glycosidase with Broad Substrate Specificity
Appl.Environ.Microbiol., 88, 2022
|
|
3PIW
 
 | | Zebrafish interferon 2 | | Descriptor: | Type I interferon 2 | | Authors: | Hamming, O.J, Hartmann, R, Lutfalla, G, Levraud, J.-P. | | Deposit date: | 2010-11-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Crystal Structure of Zebrafish Interferons I and II Reveals Conservation of Type I Interferon Structure in Vertebrates.
J.Virol., 85, 2011
|
|
7WPK
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with L-Met | | Descriptor: | METHIONINE, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPL
 
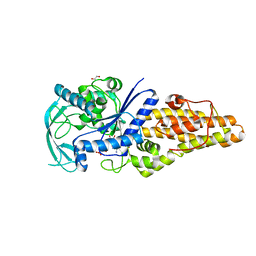 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPN
 
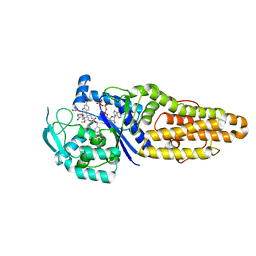 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor and ATP | | Descriptor: | (phenylmethyl) N-[(2R)-2-[2-(4-bromanyl-3-oxidanyl-phenyl)-5-(methylcarbamoyl)benzimidazol-1-yl]-2-(3,4-dimethoxyphenyl)ethyl]carbamate, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPJ
 
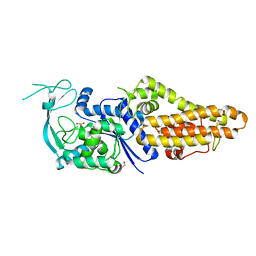 | | Methionyl-tRNA synthetase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
3O8T
 
 | |
7VE2
 
 | | Crystal Structure of Lopinavir bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, Plasmepsin II | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2021-09-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Plasmodium falciparum plasmepsins by drugs targeting HIV-1 protease: A way forward for antimalarial drug discovery.
Curr Res Struct Biol, 7, 2024
|
|
7WMC
 
 | | Crystal structure of macrocyclic peptide 1 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, Peptide1 | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
3PIV
 
 | | Zebrafish interferon 1 | | Descriptor: | Interferon, NICKEL (II) ION | | Authors: | Hamming, O.J, Hartmann, R, Lutfalla, G, Levraud, J.-P. | | Deposit date: | 2010-11-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.086 Å) | | Cite: | Crystal Structure of Zebrafish Interferons I and II Reveals Conservation of Type I Interferon Structure in Vertebrates.
J.Virol., 85, 2011
|
|
7X2Y
 
 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase in complex with NAD+ and 3-Hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, 4,5-dihydroxyphthalate dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
3Q31
 
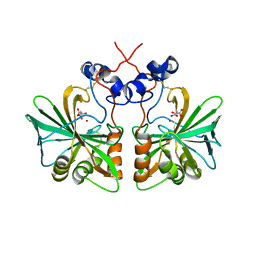 | | Structure of fungal alpha Carbonic Anhydrase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carbonic anhydrase, D-MALATE, ... | | Authors: | Cuesta-Seijo, J.A, Borchert, M.S, Navarro-Poulsen, J.C, Schnorr, K.M, Leggio, L.L. | | Deposit date: | 2010-12-21 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure of fungal alpha Carbonic Anhydrase from Aspergillus oryzae
To be Published
|
|
7W61
 
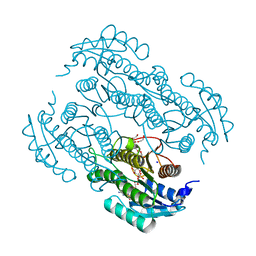 | | Crystal structure of farnesol dehydrogenase from Helicoverpa armigera | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Farnesol dehydrogenase, ... | | Authors: | Kumar, R, Das, J, Mahto, J.K, Sharma, M, Kumar, P, Sharma, A.K. | | Deposit date: | 2021-12-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and molecular characterization of NADP + -farnesol dehydrogenase from cotton bollworm, Helicoverpaarmigera.
Insect Biochem.Mol.Biol., 147, 2022
|
|
3P7F
 
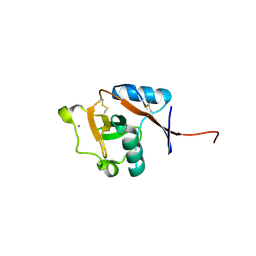 | | Structure of the human Langerin carbohydrate recognition domain | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION | | Authors: | Skerra, A, Schiefner, A. | | Deposit date: | 2010-10-12 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The carbohydrate recognition domain of Langerin reveals high structural similarity with the one of DC-SIGN but an additional, calcium-independent sugar-binding site.
Mol.Immunol., 45, 2008
|
|
