8CI9
 
 | | Deoxypodophyllotoxin Synthase in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxypodophyllotoxin synthase, FE (III) ION | | Authors: | Ingold, Z, Lichman, B, Grogan, G. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure and mutation of deoxypodophyllotoxin synthase (DPS) from Podophyllum hexandrum
Front Catal, 3, 2023
|
|
8CI8
 
 | |
8CI7
 
 | |
8CI6
 
 | | PBP AccA from A. vitis S4 in complex with D-glucose-2-phosphate (G2P) | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ABC transporter substrate binding protein (Agrocinopine) | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-08 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CHL
 
 | | Human FKBP12 in complex with (1S,5S,6R)-9-((3,5-dichlorophenyl)sulfonyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,9-diazabicyclo[4.2.1]nonan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, CADMIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHJ
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((R)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHF
 
 | | cryo-EM Structure of Craf:14-3-3:Mek1 | | Descriptor: | 14-3-3 protein zeta isoform X1, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Dedden, D, Ulrich, G. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
8CHD
 
 | | NtUGT1 in two conformations | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glycosyltransferase, ... | | Authors: | Fredslund, F, Teze, D, Adams, P.D, Welner, D.H. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | NtUGT1 in two conformations
To Be Published
|
|
8CH9
 
 | | Crystal structure of arsenite oxidase from Alcaligenes faecalis (Af Aio) bound to arsenic oxyanion | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ARSENATE, ... | | Authors: | Engrola, F, Correia, M.A.S, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of arsenite oxidase from Alcaligenes faecalis (Af Aio) bound to arsenic oxyanion
To Be Published
|
|
8CH8
 
 | | Crystal structure of the human PXR ligand-binding domain in complex with liranaftate | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, ~{O}-(5,6,7,8-tetrahydronaphthalen-2-yl) ~{N}-(6-methoxypyridin-2-yl)-~{N}-methyl-carbamothioate | | Authors: | Carivenc, C, Derosa, Q, Grimaldi, M, Boulahtouf, A, Balaguer, P, Bourguet, W. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human PXR ligand-binding domain in complex with liranaftate
To Be Published
|
|
8CH2
 
 | | PBP AccA from A. vitis S4 in complex with L-arabinose-2-phosphate (A2P) | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | Authors: | Morera, S, Deicsics, G, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CH0
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Gadolinium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Consensus tetratricopeptide repeat protein, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CGY
 
 | | Trypanosoma brucei IMP dehydrogenase (ori) crystallized in High Five cells reveals native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo; XDS processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CGV
 
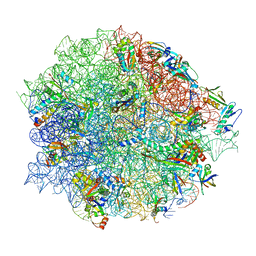 | | Tiamulin bound to the 50S subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L25, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-02-06 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.66 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGS
 
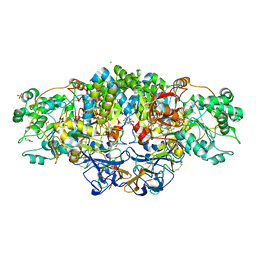 | | Crystal structure of arsenite oxidase from Alcaligenes faecalis (Af Aio) bound to antimony oxyanion | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ... | | Authors: | Engrola, F, Correia, M.A.S, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2023-02-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Arsenite oxidase in complex with antimonite and arsenite oxyanions: Insights into the catalytic mechanism.
J.Biol.Chem., 299, 2023
|
|
8CGQ
 
 | |
8CGO
 
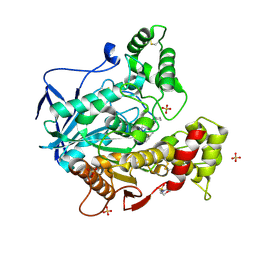 | | Structure of human butyrylcholinesterase in complex with N-{[2-(benzyloxy)-3-methoxyphenyl]methyl}-N-[3-(2-fluorophenyl)propyl]cyclobutanamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Pidany, F, Korabecny, J, Cahlikova, L, Nachon, F. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Highly selective butyrylcholinesterase inhibitors related to Amaryllidaceae alkaloids - Design, synthesis, and biological evaluation.
Eur.J.Med.Chem., 252, 2023
|
|
8CGM
 
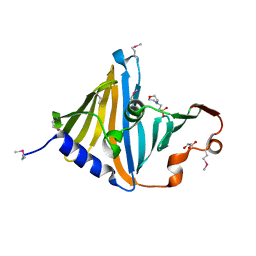 | | Structure of the lipoprotein transporter LolA from Porphyromonas gingivalis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Persson, K, Jaiman, D, Nagampalli, R. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A comparative analysis of lipoprotein transport proteins: LolA and LolB from Vibrio cholerae and LolA from Porphyromonas gingivalis.
Sci Rep, 13, 2023
|
|
8CGK
 
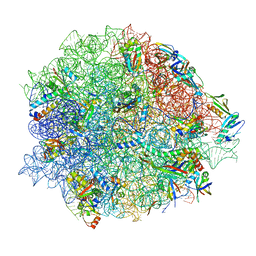 | | Lincomycin and Avilamycin bound to the 50S subunit | | Descriptor: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 23S rRNA, 50S ribosomal protein L15, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Beckert, B, Wilson, D.N. | | Deposit date: | 2023-02-05 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.64 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGC
 
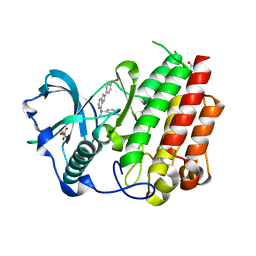 | | Structure of CSF1R in complex with a pyrollopyrimidine (compound 23) | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, Macrophage colony-stimulating factor 1 receptor, ... | | Authors: | Aarhus, T.I, Bjornstad, F, Wolowczyk, C, Larsen, K.U, Rognstad, L, Leithaug, T, Unger, A, Habenberger, P, Wolff, A, Bjorkoy, G, Pridans, C, Eickhoff, J, Klebl, B, Hoff, B.H, Sundby, E. | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Synthesis and Development of Highly Selective Pyrrolo[2,3- d ]pyrimidine CSF1R Inhibitors Targeting the Autoinhibited Form.
J.Med.Chem., 66, 2023
|
|
8CGB
 
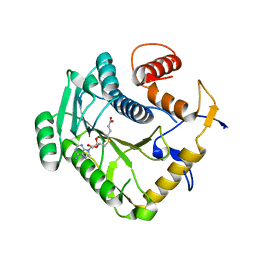 | | Monkeypox virus VP39 in complex with SAH | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Silhan, J, Klima, M, Skvara, P, Boura, E. | | Deposit date: | 2023-02-03 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CGA
 
 | | Structure of Mycobacterium tuberculosis dUTPase delta 133A-137S mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Toth, Z.S, Benedek, A, Leveles, I, Vertessy, B.G. | | Deposit date: | 2023-02-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Mycobacterium tuberculosis dUTPase delta 133A-137S mutant
To Be Published
|
|
8CG7
 
 | | Structure of p53 cancer mutant Y220C with arylation at Cys182 and Cys277 | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Balourdas, D.I, Pichon, M.M, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Reactivity Studies of 2-Sulfonylpyrimidines Allow Selective Protein Arylation.
Bioconjug.Chem., 34, 2023
|
|
8CG6
 
 | |
8CG5
 
 | |
