5TKS
 
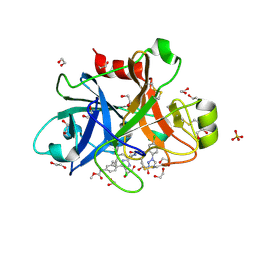 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR ((15S)-18-CHLORO- 15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2- PROPENOYL)AMINO)-17,19-DIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE | | Descriptor: | ((15S)-18-CHLORO- 15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2- PROPENOYL)AMINO)-17,19-DIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE, 1,2-ETHANEDIOL, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Design of Macrocyclic Factor XIa Inhibitors: Discovery of the Macrocyclic Amide Linker.
J. Med. Chem., 60, 2017
|
|
6E7D
 
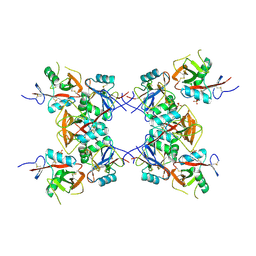 | | Structure of the inhibitory NKR-P1B receptor bound to the host-encoded ligand, Clr-b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, Killer cell lectin-like receptor subfamily B member 1B allele B, ... | | Authors: | Balaji, G.R, Rossjohn, J, Berry, R. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of host Clr-b by the inhibitory NKR-P1B receptor provides a basis for missing-self recognition.
Nat Commun, 9, 2018
|
|
5TL8
 
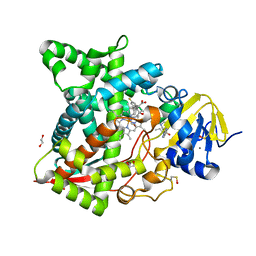 | | Naegleria fowleri CYP51-posaconazole complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, POSACONAZOLE, ... | | Authors: | Podust, L.M, Jennings, G, Calvet-Alvarez, C, Debnath, A. | | Deposit date: | 2016-10-10 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of the Naegleria fowleri CYP51 at 1.7 Angstroms resolution
To be published
|
|
8TXG
 
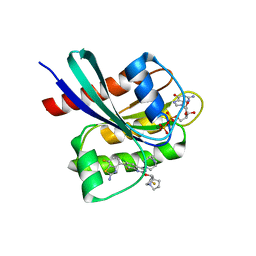 | | Crystal structure of KRAS G12D in complex with GDP and compound 8 | | Descriptor: | (4M)-4-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-7-fluoro-1,3-benzothiazol-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
6RO8
 
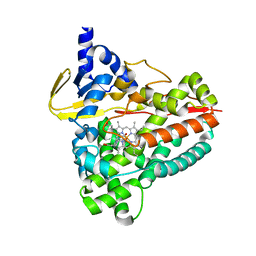 | | The crystal structure of Acinetobacter radioresistens CYP116B5 heme domain | | Descriptor: | Cytochrome P450 RhF, HISTIDINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ciaramella, A, Catucci, G, Gilardi, G, Di Nardo, G. | | Deposit date: | 2019-05-10 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of bacterial CYP116B5 heme domain: New insights on class VII P450s structural flexibility and peroxygenase activity.
Int.J.Biol.Macromol., 140, 2019
|
|
6E9A
 
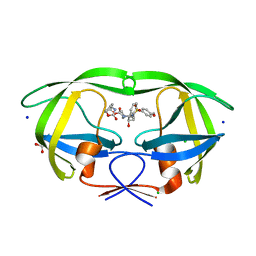 | | HIV-1 WILD TYPE PROTEASE WITH GRL-034-17A, (3aS, 5R, 6aR)-2-OXOHEXAHYD CYCLOPENTA[D]-5-OXAZOLYL URETHANE WITH A BICYCLIC OXAZOLIDINONE SCAFF AS THE P2 LIGAND | | Descriptor: | (3aS,5R,6aR)-2-oxohexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
4XH6
 
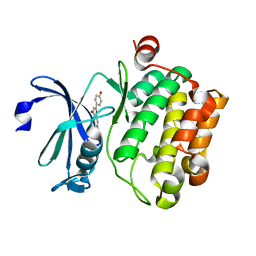 | | Crystal structure of proto-oncogene kinase Pim1 bound to hispidulin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-4H-chromen-4-one, Serine/threonine-protein kinase pim-1 | | Authors: | Su, M.-Y, Chang, C.-I. | | Deposit date: | 2015-01-05 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Total Synthesis of Hispidulin and the Structural Basis for Its Inhibition of Proto-oncogene Kinase Pim-1
J.Nat.Prod., 78, 2015
|
|
6ROD
 
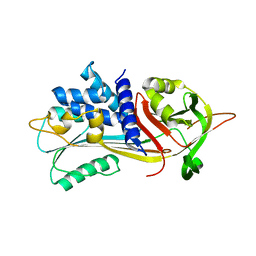 | |
6ROL
 
 | | Structure of IMP2 KH34 domains | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Insulin-like growth factor 2 mRNA-binding protein 2, ... | | Authors: | Bhaskar, V, Biswas, J, Singer, R.H, Chao, J.A. | | Deposit date: | 2019-05-13 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for RNA selectivity by the IMP family of RNA-binding proteins.
Nat Commun, 10, 2019
|
|
6E9W
 
 | | Crystal structure of Rock1 with a pyridinylbenzamide based inhibitor | | Descriptor: | N-[(2,3-dihydro-1,4-benzodioxin-5-yl)methyl]-4-(pyridin-4-yl)benzamide, Rho-associated protein kinase 1, SULFATE ION | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification of Selective Dual ROCK1 and ROCK2 Inhibitors Using Structure Based Drug Design.
J. Med. Chem., 61, 2018
|
|
5TRB
 
 | | Crystal structure of the RNF20 RING domain | | Descriptor: | E3 ubiquitin-protein ligase BRE1A, ZINC ION | | Authors: | Foglizzo, M, Middleton, A.J, Day, C.L. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the RING Domains of RNF20 and RNF40, Dimeric E3 Ligases that Monoubiquitylate Histone H2B.
J.Mol.Biol., 428, 2016
|
|
8TXH
 
 | | Crystal structure of KRAS G12D in complex with GDP and compound 14 | | Descriptor: | (4P)-2-amino-4-{4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}-6-(trifluoromethyl)quinazolin-7-yl}-7-fluoro-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
6RQ1
 
 | | CYP121 in complex with 2-methyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(2-methyl-4-oxidanyl-phenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
4OEI
 
 | | Crystal structure of plant lectin from Cicer arietinum at 2.6 angstrom resolution | | Descriptor: | Lectin, MAGNESIUM ION, SULFATE ION | | Authors: | Kumar, S, Dube, D, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-13 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of plant lectin from Cicer arietinum at
2.6 angstrom resolution
TO BE PUBLISHED
|
|
6RQB
 
 | | CYP121 in complex with 3-bromo dicyclotyrosine | | Descriptor: | 3-bromo dicyclotyrosine, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQ9
 
 | | CYP121 in complex with O-methyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(4-methoxyphenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
4OFF
 
 | |
7RPZ
 
 | | KRAS G12D in complex with MRTX-1133 | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)-5-ethynyl-6-fluoronaphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-05 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
4OFG
 
 | | Co-crystal structure of carboxy cGMP binding domain of Plasmodium falciparum PKG with cGMP | | Descriptor: | CGMP-dependent protein kinase, CYCLIC GUANOSINE MONOPHOSPHATE, ETHANOL, ... | | Authors: | Kim, J.J, Sanabria figueroa, E, Kim, C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Carboxyl cGMP Binding Domain of the Plasmodium falciparum cGMP-dependent Protein Kinase Reveal a Novel Capping Triad Crucial for Merozoite Egress.
Plos Pathog., 11, 2015
|
|
7R9L
 
 | | Crystal structure of HPK1 in complex with compound 2 | | Descriptor: | 2-amino-N,N-dimethyl-5-(1H-pyrrolo[2,3-b]pyridin-5-yl)benzamide, Hematopoietic progenitor kinase | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
8TXE
 
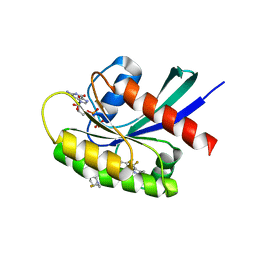 | | Crystal structure of KRAS G12D in complex with GDP and compound 5 | | Descriptor: | (6M)-6-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-4-methyl-5-(trifluoromethyl)pyridin-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
6EGZ
 
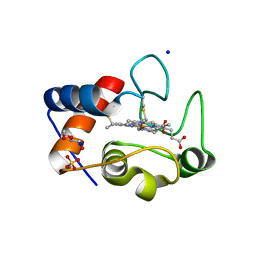 | | Crystal structure of cytochrome c in complex with di-PEGylated sulfonatocalix[4]arene | | Descriptor: | Cytochrome c iso-1, HEME C, SODIUM ION, ... | | Authors: | Mummidivarapu, V.V.S, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Noncovalent PEGylation via Sulfonatocalix[4]arene-A Crystallographic Proof.
Bioconjug.Chem., 29, 2018
|
|
7R9P
 
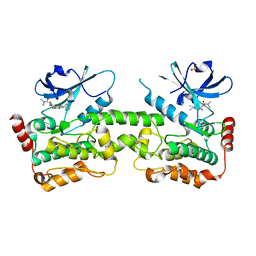 | | Crystal structure of HPK1 in complex with compound 14 | | Descriptor: | 6-amino-2-fluoro-N,N-dimethyl-3-(4'-methylspiro[cyclopropane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl)benzamide, Hematopoietic progenitor kinase, SULFATE ION | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
6RUH
 
 | |
5U6W
 
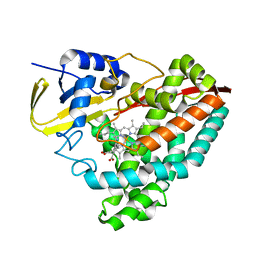 | | The crystal structure of 4-methylaminobenzoate-bound CYP199A4 | | Descriptor: | 4-(methylamino)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Coleman, T, Bruning, J.B, Bell, S.G. | | Deposit date: | 2016-12-09 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Cytochrome P450 CYP199A4 from Rhodopseudomonas palustris Catalyzes Heteroatom Dealkylations, Sulfoxidation, and Amide and Cyclic Hemiacetal Formation
Acs Catalysis, 8, 2018
|
|
