7GJQ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-78e1d523-1 (Mpro-P0627) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzothiopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3KZE
 
 | |
7GTM
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000543a | | Descriptor: | (4S)-4-hydroxy-2-(propan-2-yl)-3,4-dihydro-1lambda~6~,2-benzothiazine-1,1(2H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
3L58
 
 | | Structure of BACE Bound to SCH589432 | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
7GTK
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000552a | | Descriptor: | (4R)-2-(2-hydroxyethyl)-4-methoxy-3,4-dihydro-1lambda~6~,2-benzothiazine-1,1(2H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
2YXO
 
 | | Histidinol Phosphate Phosphatase complexed with Sulfate | | Descriptor: | FE (III) ION, GLYCEROL, Histidinol phosphatase, ... | | Authors: | Omi, R. | | Deposit date: | 2007-04-26 | | Release date: | 2007-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Monofunctional Histidinol Phosphate Phosphatase from Thermus thermophilus HB8.
Biochemistry, 46, 2007
|
|
5TFV
 
 | |
4DBC
 
 | | Substrate Activation in Aspartate Aminotransferase | | Descriptor: | (E)-N-{2-hydroxy-3-methyl-6-[(phosphonooxy)methyl]benzylidene}-L-aspartic acid, 1,2-ETHANEDIOL, Aspartate aminotransferase, ... | | Authors: | Toney, M.D, Fisher, A.J, Griswold, W.R. | | Deposit date: | 2012-01-14 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ground-state electronic destabilization via hyperconjugation in aspartate aminotransferase.
J.Am.Chem.Soc., 134, 2012
|
|
5RYF
 
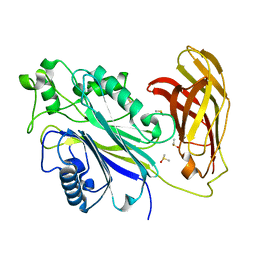 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1266823232 | | Descriptor: | (1S)-1-(2,4-dimethyl-1,3-thiazol-5-yl)-N-methylethan-1-amine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5TM7
 
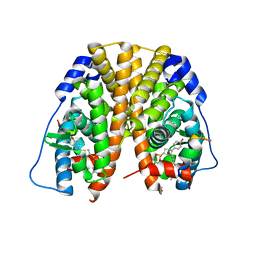 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 7-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)heptanoic acid | | Descriptor: | 7-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}heptanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
3LEA
 
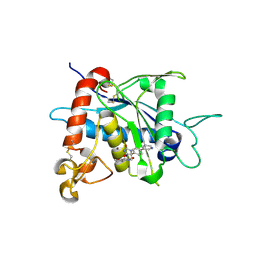 | |
7GIJ
 
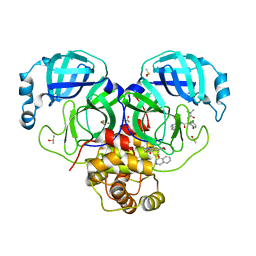 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-5d65ec79-1 (Mpro-P0097) | | Descriptor: | (4S)-6-chloro-4-[2-(dimethylamino)-2-oxoethyl]-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5UPG
 
 | |
5ZQK
 
 | | Dengue Virus Non Structural Protein 5 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | El Sahili, A, Soh, T.S, Schiltz, J, Gharbi-Ayachi, A, Goh, B.C, Seh, C.C, Dedon, P.C, Shi, P.Y, Lim, S.P, Lescar, J. | | Deposit date: | 2018-04-19 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NS5 from Dengue Virus Serotype 2 Can Adopt a Conformation Analogous to That of Its Zika Virus and Japanese Encephalitis Virus Homologues.
J.Virol., 94, 2019
|
|
5RZ7
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z57472297 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-methyl-1,3-bis(oxidanyl)propan-2-yl]-3-phenyl-urea, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
2YCP
 
 | | F448H mutant of tyrosine phenol-lyase from Citrobacter freundii in complex with quinonoid intermediate formed with 3-fluoro-L-tyrosine | | Descriptor: | (2E)-3-(3-fluoro-4-hydroxyphenyl)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}propanoic acid, 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Milic, D, Demidkina, T.V, Faleev, N.G, Phillips, R.S, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Snapshots of Tyrosine Phenol-Lyase Show that Substrate Strain Plays a Role in C-C Bond Cleavage
J.Am.Chem.Soc., 133, 2011
|
|
3DKL
 
 | | Crystal structure of phosphorylated mimic form of human NAMPT complexed with benzamide and phosphoribosyl pyrophosphate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, BENZAMIDE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ho, M, Burgos, E.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-06-25 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DYB
 
 | | proteinase K- digalacturonic acid complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Proteinase K, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A, Cudney, R, Nguyen, C, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | High-resolution structure of proteinase K cocrystallized with digalacturonic acid.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5ZFL
 
 | | Crystal structure of beta-lactamase PenP mutant E166Y | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase | | Authors: | Pan, X, Zhao, Y. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The hydrolytic water molecule of Class A beta-lactamase relies on the acyl-enzyme intermediate ES* for proper coordination and catalysis.
Sci Rep, 10, 2020
|
|
2Y8E
 
 | |
3LSW
 
 | | Aniracetam bound to the ligand binding domain of GluA3 | | Descriptor: | 1-(4-METHOXYBENZOYL)-2-PYRROLIDINONE, GLUTAMIC ACID, GluA2 S1S2 domain, ... | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2010-02-13 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Piracetam Defines a New Binding Site for Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazole-propionic Acid (AMPA) Receptors.
J.Med.Chem., 53, 2010
|
|
5R8B
 
 | | PanDDA analysis group deposition INTERLEUKIN-1 BETA -- Fragment Z2027049478 in complex with INTERLEUKIN-1 BETA | | Descriptor: | 5-(1,3-thiazol-2-yl)-1H-1,2,4-triazole, Interleukin-1 beta, SULFATE ION | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
5UT0
 
 | | JAK2 JH2 in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Puleo, D.E, Schlessinger, J. | | Deposit date: | 2017-02-14 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Identification and Characterization of JAK2 Pseudokinase Domain Small Molecule Binders.
ACS Med Chem Lett, 8, 2017
|
|
7GPK
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z905434478 | | Descriptor: | 1-[(thiophen-3-yl)methyl]piperidin-4-ol, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
3L7G
 
 | | Crystal structure of organophosphate anhydrolase/prolidase | | Descriptor: | MANGANESE (II) ION, N,N'-bis(1-methylethyl)phosphorodiamidic acid, Xaa-Pro dipeptidase | | Authors: | Vyas, N.K, Nickitenko, A, Quiocho, F.A. | | Deposit date: | 2009-12-28 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the dual activities of the nerve agent degrading organophosphate anhydrolase/prolidase.
Biochemistry, 49, 2010
|
|
