1UR9
 
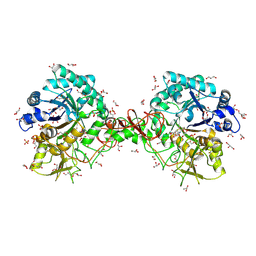 | | Interactions of a family 18 chitinase with the designed inhibitor HM508, and its degradation product, chitobiono-delta-lactone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, CHITINASE B, GLYCEROL, ... | | Authors: | Vaaje-Kolstad, G, Vasella, A, Peter, M.G, Netter, C, Houston, D.R, Westereng, B, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2003-10-27 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of a Family 18 Chitinase with the Designed Inhibitor Hm508 and its Degradation Product, Chitobiono-Delta-Lactone.
J.Biol.Chem., 279, 2004
|
|
1VC6
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Product with C75U Mutaion, cleaved in Imidazole and Mg2+ solutions | | Descriptor: | Hepatitis Delta virus ribozyme, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1HJX
 
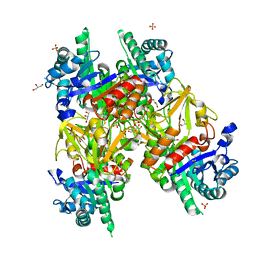 | | Ligand-induced signalling and conformational change of the 39 kD glycoprotein from human articular chondrocytes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE-3 LIKE PROTEIN 1, GLYCEROL, ... | | Authors: | Houston, D.R, Recklies, A.D, Krupa, J.C, Van Aalten, D.M.F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Ligand-Induced Conformational Change of the 39-kDa Glycoprotein from Human Articular Chondrocytes
J.Biol.Chem., 278, 2003
|
|
1HN6
 
 | |
1V1P
 
 | |
1VBX
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in EDTA solution | | Descriptor: | Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
2KQH
 
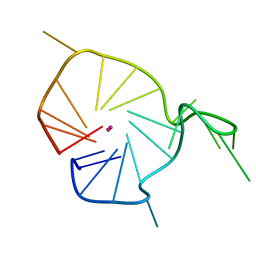 | | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*GP*GP*GP*CP*GP*CP*GP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3', POTASSIUM ION | | Authors: | Hsu, S.-T.D, Varnai, P, Bugaut, A, Reszka, A.P, Neidle, S, Balasubramanian, S. | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics
J.Am.Chem.Soc., 131, 2009
|
|
1US2
 
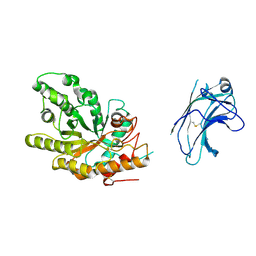 | | Xylanase10C (mutant E385A) from Cellvibrio japonicus in complex with xylopentaose | | Descriptor: | ENDO-BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
1HX5
 
 | |
1HTB
 
 | |
1VBZ
 
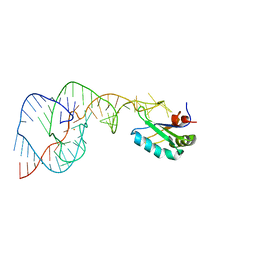 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Ba2+ solution | | Descriptor: | BARIUM ION, Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VC5
 
 | | Crystal Structure of the Wild Type Hepatitis Delta Virus Gemonic Ribozyme Precursor, in EDTA solution | | Descriptor: | Hepatitis Delta virus ribozyme, SODIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1UWX
 
 | | P1.2 serosubtype antigen derived from N. meningitidis PorA in complex with Fab fragment | | Descriptor: | ANTIBODY, CLASS 1 OUTER MEMBRANE PROTEIN VARIABLE REGION 2, PROTEIN G-PRIME | | Authors: | Tzitzilonis, C, Prince, S.M, Collins, R.F, Maiden, M.C.J, Feavers, I.M, Derrick, J.P. | | Deposit date: | 2004-02-12 | | Release date: | 2005-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Variation and Immune Recognition of the P1.2 Subtype Meningococcal Antigen.
Proteins: Struct., Funct., Bioinf., 62, 2005
|
|
1UOM
 
 | | The Structure of Estrogen Receptor in Complex with a Selective and Potent Tetrahydroisochiolin Ligand. | | Descriptor: | 2-PHENYL-1-[4-(2-PIPERIDIN-1-YL-ETHOXY)-PHENYL]-1,2,3,4-TETRAHYDRO-ISOQUINOLIN-6-OL, ESTROGEN RECEPTOR | | Authors: | Stark, W, Bischoff, S.F, Buhl, T, Fournier, B, Halleux, C, Kallen, J, Keller, H, Renaud, J. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Estrogen Receptor Modulators: Identification and Structure-Activity Relationships of Potent Eralpha-Selective Tetrahydroisoquinoline Ligands
J.Med.Chem., 46, 2003
|
|
1UUZ
 
 | | IVY:A NEW FAMILY OF PROTEIN | | Descriptor: | INHIBITOR OF VERTEBRATE LYSOZYME, LYSOZYME C | | Authors: | Abergel, C, Lembo, F, Byrne, D, Maza, C, Claverie, J.M. | | Deposit date: | 2004-01-12 | | Release date: | 2004-01-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Evolution of the Ivy Protein Family, Unexpected Lysozyme Inhibitors in Gram-Negative Bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1HSQ
 
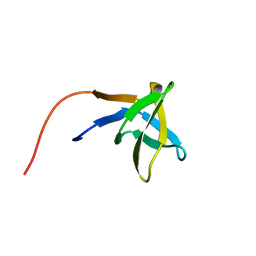 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
1HTO
 
 | |
1HPT
 
 | |
1HJW
 
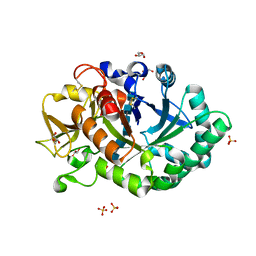 | | Crystal structure of hcgp-39 in complex with chitin octamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Houston, D.R, Recklies, A.D, Krupa, J.C, Van Aalten, D.M.F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Ligand-Induced Conformational Change of the 39-kDa Glycoprotein from Human Articular Chondrocytes
J.Biol.Chem., 278, 2003
|
|
1TGB
 
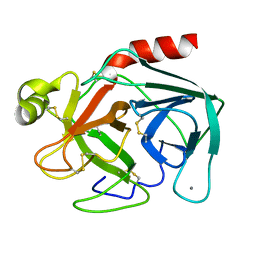 | | CRYSTAL STRUCTURE OF BOVINE TRYPSINOGEN AT 1.8 ANGSTROMS RESOLUTION. II. CRYSTALLOGRAPHIC REFINEMENT, REFINED CRYSTAL STRUCTURE AND COMPARISON WITH BOVINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSINOGEN | | Authors: | Bode, W, Fehlhammer, H, Huber, R. | | Deposit date: | 1979-03-07 | | Release date: | 1979-06-13 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of bovine trypsinogen at 1-8 A resolution. II. Crystallographic refinement, refined crystal structure and comparison with bovine trypsin.
J.Mol.Biol., 111, 1977
|
|
1SRU
 
 | | Crystal structure of full length E. coli SSB protein | | Descriptor: | Single-strand binding protein | | Authors: | Savvides, S.N, Raghunathan, S, Fuetterer, K, Kozlov, A.G, Lohman, T.M, Waksman, G. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA.
Protein Sci., 13, 2004
|
|
2KOG
 
 | | lipid-bound synaptobrevin solution NMR structure | | Descriptor: | Vesicle-associated membrane protein 2 | | Authors: | Ellena, J.F, Liang, B, Wiktor, M, Stein, A, Cafiso, D.S, Jahn, R, Tamm, L.K. | | Deposit date: | 2009-09-22 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KTR
 
 | | NMR structure of p62 PB1 dimer determined based on PCS | | Descriptor: | Sequestosome-1, TERBIUM(III) ION | | Authors: | Saio, T, Yokochi, M, Kumeta, H, Inagaki, F. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | PCS-based structure determination of protein-protein complexes
J.Biomol.Nmr, 46, 2010
|
|
1V1O
 
 | |
2KPM
 
 | | Solution NMR Structure of uncharacterized protein from gene locus NE0665 of Nitrosomonas europaea. Northeast Structural Genomics Target NeR103A | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Belote, R, Jiang, M, Xiao, R, Ciccosanti, C, Acton, T, Everett, J, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of uncharacterized protein from gene locus NE0665 of Nitrosomonas europaea. Northeast Structural Genomics Target NeR103A
To be Published
|
|
