8Q63
 
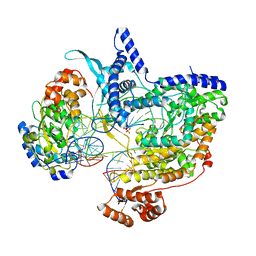 | | Cryo-EM structure of IC8', a second state of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-08-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
4ZN3
 
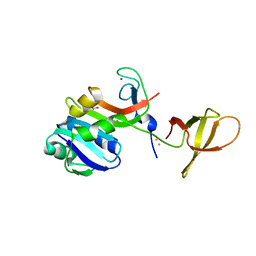 | | Crystal Structure of MjSpt4:Spt5 complex conformation B | | Descriptor: | FE (III) ION, GLYCEROL, Transcription elongation factor Spt4, ... | | Authors: | Guo, G.R, Zhou, H.H, Gao, Y.X, Zhu, Z.L, Niu, L.W, Teng, M.K. | | Deposit date: | 2015-05-04 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical insights into the DNA-binding mode of MjSpt4p:Spt5 complex at the exit tunnel of RNAPII
J.Struct.Biol., 192, 2015
|
|
8AP1
 
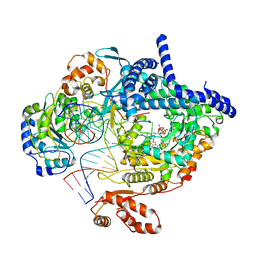 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with two GTP molecules poised for de novo initiation (IC2) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATV
 
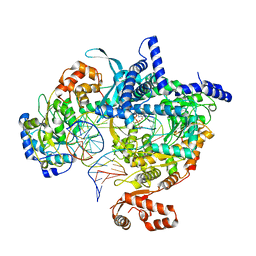 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 5-mer RNA, pppGpGpApApA (IC5) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATW
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 6-mer RNA, pppGpGpApApApU (IC6) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATT
 
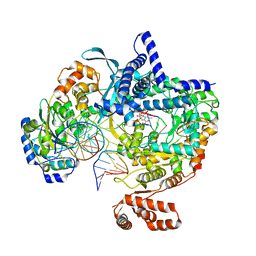 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 4-mer RNA, pppGpGpUpA (IC4) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
7Y9Y
 
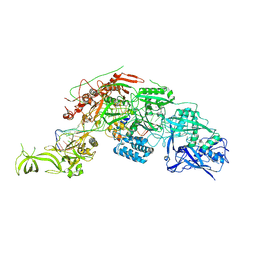 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (no PFS) complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, RNA (27-MER), ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7Y9X
 
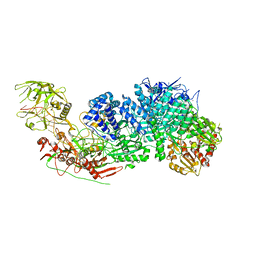 | | Structure of the Cas7-11-Csx29-guide RNA complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, ZINC ION, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
1AJF
 
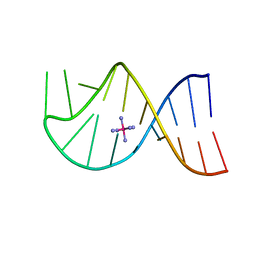 | |
5NOH
 
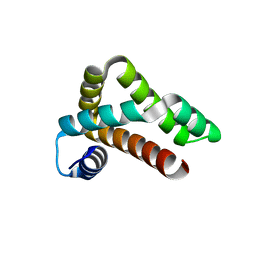 | | HRSV M2-1 core domain | | Descriptor: | Transcription elongation factor M2-1 | | Authors: | Josts, I, Almeida Hernandez, Y, Molina, I.G, de Prat-Gay, G, Tidow, H. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the Human respiratory syncytial virus M2-1RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5NKX
 
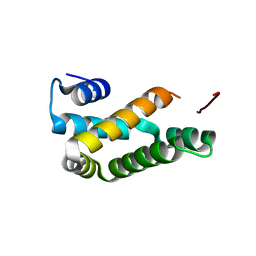 | | HRSV M2-1 core domain, P3221 crystal form | | Descriptor: | M2-1 | | Authors: | Almeida Hernandez, Y, Josts, I, Molina, I.G, de Pray-Gay, G, Tidow, H. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.00008678 Å) | | Cite: | Structure and stability of the Human respiratory syncytial virus M2-1RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1XSU
 
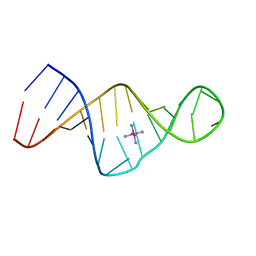 | |
1XST
 
 | |
2VOO
 
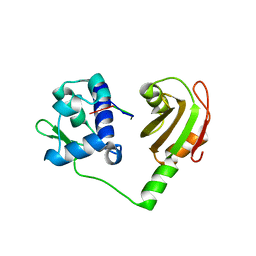 | | Crystal structure of N-terminal domains of Human La protein complexed with RNA oligomer UUUUUUUU | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP)-3', LUPUS LA PROTEIN | | Authors: | Kotik-Kogan, O, Valentine, E.R, Sanfelice, D, Conte, M.R, Curry, S. | | Deposit date: | 2008-02-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis Reveals Conformational Plasticity in the Recognition of RNA 3' Ends by the Human La Protein.
Structure, 16, 2008
|
|
5HCI
 
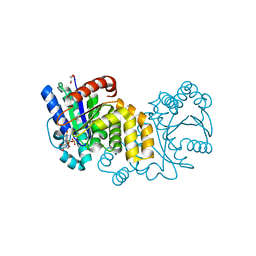 | | GPN-loop GTPase Npa3 in complex with GDP | | Descriptor: | GLYCEROL, GPN-loop GTPase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niesser, J, Wagner, F.R, Kostrewa, D, Muehlbacher, W, Cramer, P. | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly.
Mol.Cell.Biol., 36, 2015
|
|
5HCN
 
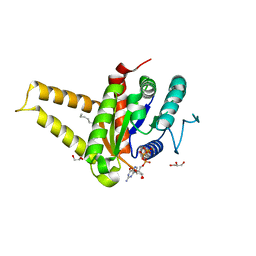 | | GPN-loop GTPase Npa3 in complex with GMPPCP | | Descriptor: | GLYCEROL, GPN-loop GTPase 1, LAURIC ACID, ... | | Authors: | Niesser, J, Wagner, F.R, Kostrewa, D, Muehlbacher, W, Cramer, P. | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly.
Mol.Cell.Biol., 36, 2015
|
|
4PUF
 
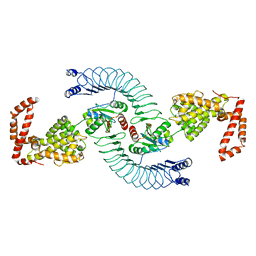 | | Complex between the Salmonella T3SS effector SlrP and its human target thioredoxin-1 | | Descriptor: | E3 ubiquitin-protein ligase SlrP, Thioredoxin | | Authors: | Zouhir, S, Bernal-Bayard, J, Cordero-Alba, M, Cardenal-Munoz, E, Guimaraes, B, Lazar, N, Ramos-Morales, F, Nessler, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | The structure of the Slrp-Trx1 complex sheds light on the autoinhibition mechanism of the type III secretion system effectors of the NEL family.
Biochem.J., 464, 2014
|
|
1NGM
 
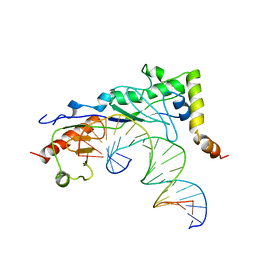 | | Crystal structure of a yeast Brf1-TBP-DNA ternary complex | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*CP*AP*TP*TP*TP*TP*TP*TP*TP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*AP*AP*AP*AP*AP*AP*AP*TP*GP*TP*TP*TP*TP*TP*T)-3', Transcription factor IIIB BRF1 subunit, ... | | Authors: | Juo, Z.S, Kassavetis, G.A, Wang, J, Geiduschek, E.P, Sigler, P.B. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a transcription factor IIIB core interface ternary complex
Nature, 422, 2003
|
|
8TDZ
 
 | |
2VON
 
 | | Crystal structure of N-terminal domains of Human La protein complexed with RNA oligomer AUAAUUU | | Descriptor: | 5'-R(*AP*UP*AP*AP*UP*UP*UP)-3', LUPUS LA PROTEIN | | Authors: | Kotik-Kogan, O, Valentine, E.R, Sanfelice, D, Conte, M.R, Curry, S. | | Deposit date: | 2008-02-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis Reveals Conformational Plasticity in the Recognition of RNA 3' Ends by the Human La Protein.
Structure, 16, 2008
|
|
5UI6
 
 | | Solution NMR Structure of Lasso Peptide Acinetodin | | Descriptor: | Acinetodin | | Authors: | Bushin, L.B, Metelev, M, Severinov, K, Seyedsayamdost, M.R. | | Deposit date: | 2017-01-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
ACS Chem. Biol., 12, 2017
|
|
5UI7
 
 | | Solution NMR Structure of Lasso Peptide Klebsidin | | Descriptor: | Klebsidin | | Authors: | Bushin, L.B, Metelev, M, Severinov, K, Seyedsayamdost, M.R. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
ACS Chem. Biol., 12, 2017
|
|
1TF6
 
 | | CO-CRYSTAL STRUCTURE OF XENOPUS TFIIIA ZINC FINGER DOMAIN BOUND TO THE 5S RIBOSOMAL RNA GENE INTERNAL CONTROL REGION | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*GP*CP*CP*TP*GP*GP*TP*TP*AP*GP*TP*AP*C P*CP*TP*GP*GP*AP* TP*GP*GP*GP*AP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*TP*CP*CP*CP*AP*TP*CP*CP*AP*GP*GP*T P*AP*CP*TP*AP*AP* CP*CP*AP*GP*GP*CP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR IIIA), ... | | Authors: | Nolte, R.T, Conlin, R.M, Harrison, S.C, Brown, R.S. | | Deposit date: | 1998-03-02 | | Release date: | 1998-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6IVU
 
 | |
8B3G
 
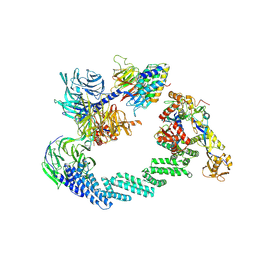 | | C(N)RL4CSA-UVSSA-E2-ubiquitin complex. | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for RNA polymerase II ubiquitylation and inactivation in transcription-coupled repair.
Nat.Struct.Mol.Biol., 31, 2024
|
|
