6CNZ
 
 | |
4S1G
 
 | |
2XV0
 
 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAHAAM), chemically reduced, pH4.8 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
4BUW
 
 | | Crystal structure of human tankyrase 2 in complex with 2-(4-(2-oxo-1, 3-oxazolidin-3-yl)phenyl)-3,4-dihydroquinazolin-4-one | | Descriptor: | 2-[4-(2-OXO-1,3-OXAZOLIDIN-3-YL)PHENYL]-3,4-DIHYDROQUINAZOLIN-4-ONE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
5U4F
 
 | | Wild-type Transthyretin in complex with 1,1'-(1E)-(1,2-Ethenediyl)bis[2-chloro-4-boronic acid]benzene | | Descriptor: | Transthyretin, [(E)-ethene-1,2-diylbis(3-chloro-4,1-phenylene)]diboronic acid | | Authors: | Windsor, I.W, Smith, T.P, Raines, R.T, Forest, K.T. | | Deposit date: | 2016-12-03 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stilbene Boronic Acids Form a Covalent Bond with Human Transthyretin and Inhibit Its Aggregation.
J. Med. Chem., 60, 2017
|
|
5TYG
 
 | | DNA Polymerase Mu Product Complex, 10 mM Mg2+ (960 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-19 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
5DPG
 
 | | sfGFP mutant - 133 p-cyano-L-phenylalanine | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, SODIUM ION | | Authors: | Dippel, A.B, Olenginski, G.M, Maurici, N, Liskov, M.T, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2015-09-12 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the effectiveness of spectroscopic reporter unnatural amino acids: a structural study.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5TYU
 
 | | DNA Polymerase Mu Reactant Complex, Mn2+ (4 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-08-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
7GE2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-6af13d92-1 (Mpro-x11313) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[2-(2-methoxyphenoxy)ethyl]-5-methyl-2-oxo-1,2-dihydroquinoline-4-carboxamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2XSD
 
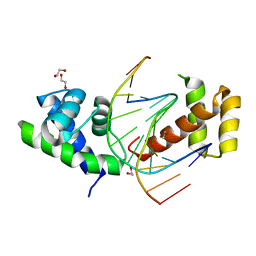 | | Crystal Structure of the dimeric Oct-6 (Pou3f1) POU domain bound to palindromic MORE DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*TP*GP*CP*AP*TP*GP*AP*GP*GP*AP)-3', 5'-D(*CP*CP*TP*CP*AP*TP*GP*CP*AP*TP*AP)-3', ... | | Authors: | Jauch, R, Choo, S.H, Ng, C.K.L, Kolatkar, P.R. | | Deposit date: | 2010-09-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Crystal Structure of the Dimeric Oct6 (POU3F1) POU Domain Bound to Palindromic More DNA.
Proteins, 79, 2011
|
|
5YUI
 
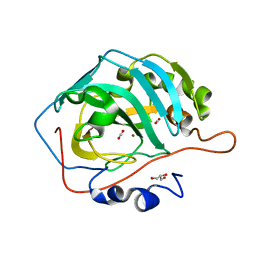 | | CO2 release in human carbonic anhydrase II crystals: reveal histidine 64 and solvent dynamics | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Park, S.Y, McKenna, R. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Tracking solvent and protein movement during CO2 release in carbonic anhydrase II crystals.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6N45
 
 | |
2XGE
 
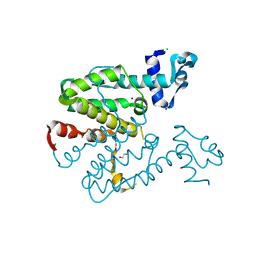 | | Crystal structure of a designed heterodimeric variant T-A(A)B of the tetracycline repressor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Stiebritz, M.T, Wengrzik, S, Richter, J.P, Muller, Y.A. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Computational Design of a Chain-Specific Tetracycline Repressor Heterodimer.
J.Mol.Biol., 403, 2010
|
|
3KYD
 
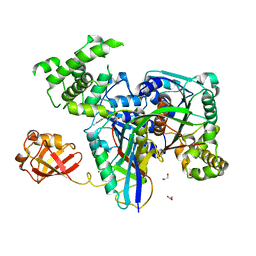 | | Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic | | Descriptor: | 1,2-ETHANEDIOL, 5'-{[(3-aminopropyl)sulfonyl]amino}-5'-deoxyadenosine, SUMO-activating enzyme subunit 1, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
1ZPG
 
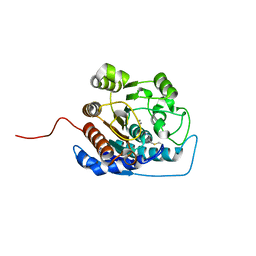 | | Arginase I covalently modified with propylamine at Q19C | | Descriptor: | Arginase 1, MANGANESE (II) ION | | Authors: | Colleluori, D.M, Reczkowski, R.S, Emig, F.A, Cama, E, Cox, J.D, Scolnick, L.R, Compher, K, Jude, K, Han, S, Viola, R.E, Christianson, D.W, Ash, D.E. | | Deposit date: | 2005-05-16 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
7GCU
 
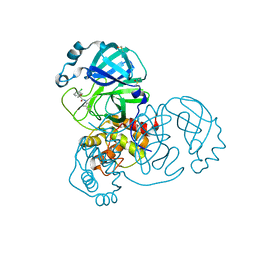 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-0f94fc3d-25 (Mpro-x10787) | | Descriptor: | 1-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)-3-oxocyclobutane-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6VGZ
 
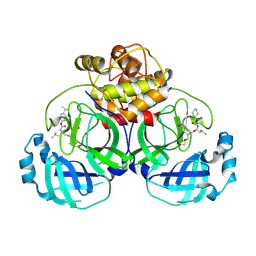 | | 2.25 A resolution structure of MERS 3CL protease in complex with inhibitor 6d | | Descriptor: | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[trans-4-(propan-2-yl)cyclohexyl]oxy}carbonyl)-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
3L12
 
 | |
7GEZ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-6747fa38-1 (Mpro-x11541) | | Descriptor: | 2-(4-acetylpiperazin-1-yl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4RUL
 
 | | Crystal structure of full-length E.Coli topoisomerase I in complex with ssDNA | | Descriptor: | DNA topoisomerase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Chen, B, Tse-Dinh, Y.C. | | Deposit date: | 2014-11-20 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I.
Nucleic Acids Res., 43, 2015
|
|
5DMR
 
 | |
9KBT
 
 | |
4RQ9
 
 | | Crystal structure of the chromophore-binding domain of Stigmatella aurantiaca bacteriophytochrome (Thr289His mutant) in the Pr state | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, GLYCEROL, ... | | Authors: | Woitowich, N.C, Halavaty, A.S, Gallagher, K.D, Nugent, A.C, Patel, H, Duong, P, Kovaleva, S.E, St.Peter, S, Ozarowski, W.B, Hernandez, C.N, Stojkovic, E.A. | | Deposit date: | 2014-10-31 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the chromophore-binding domain of Stigmatella aurantiaca bacteriophytochrome (Thr289His mutant) in the Pr state
To be Published
|
|
5TXZ
 
 | | DNA Polymerase Mu Reactant Complex, 100mM Mg2+ (15 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-17 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
7GGR
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-c7771779-1 (Mpro-x12695) | | Descriptor: | (4S)-6-chloro-4-hydroxy-N-(isoquinolin-4-yl)-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
