5D6R
 
 | |
5DEX
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, phenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-phenyl-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 2017
|
|
3JQL
 
 | | Crystal Structure of the Complex Formed Between Phospholipase A2 and a Hexapeptide Fragment of Amyloid Beta Peptide, Lys-Leu-Val-Phe-Phe-Ala at 1.2 A Resolution | | Descriptor: | Acidic phospholipase A2 3 (Fragment), Amyloid Beta Peptide, CALCIUM ION | | Authors: | Mirza, Z, Vikram, G, Singh, N, Sinha, M, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2009-09-07 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of the Complex Formed Between Phospholipase A2 and a Hexapeptide Fragment of Amyloid Beta Peptide, Lys-Leu-Val-Phe-Phe-Ala at 1.2 A Resolution
To be Published
|
|
7B21
 
 | | The X183 domain from Cellvibrio japonicus Cbp2D | | Descriptor: | 1,2-ETHANEDIOL, Carbohydrate binding protein, putative, ... | | Authors: | Branch, J, Hemsworth, G.R. | | Deposit date: | 2020-11-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | C-type cytochrome-initiated reduction of bacterial lytic polysaccharide monooxygenases.
Biochem.J., 478, 2021
|
|
7KIC
 
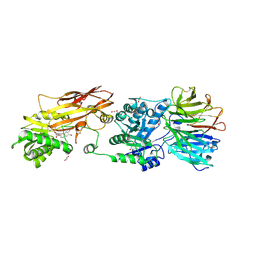 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 34 | | Descriptor: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, Methylosome protein 50, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
5POE
 
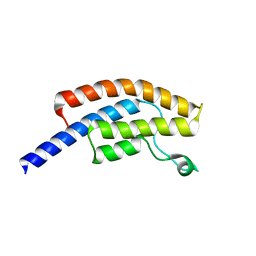 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10188a and N07807b | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4-phenyl-5-(1H-pyrrol-1-yl)-1H-pyrazole, 6-amino-1-methylquinolin-2(1H)-one, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.518 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5POQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10974a | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(methylsulfonyl)piperazin-1-yl]ethan-1-one, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
155L
 
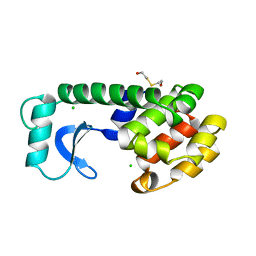 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
164L
 
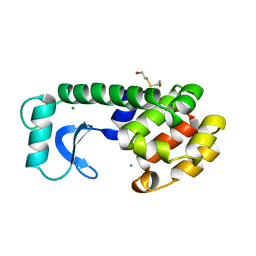 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
6M80
 
 | | Collagen peptide containing aza-proline and aza-glycine | | Descriptor: | 1,2-ETHANEDIOL, Collagen peptide containing aza-proline and aza-glycine, SULFATE ION | | Authors: | Chenoweth, D.M, Kasznel, A.J, Porter, N.J. | | Deposit date: | 2018-08-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Aza-proline effectively mimics l-proline stereochemistry in triple helical collagen.
Chem Sci, 10, 2019
|
|
4P8W
 
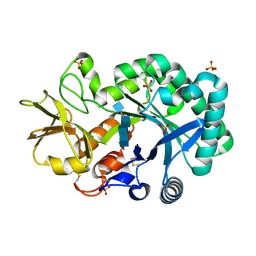 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc4) were solved to resolutions of 1.9 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
7L9Y
 
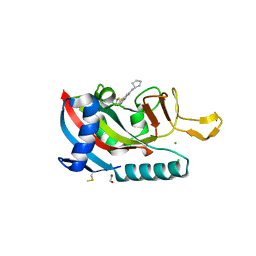 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN012042 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopentylamino)-5-fluoro-2-{[(piperidin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, CHLORIDE ION, ... | | Authors: | Dorsey, B.W, Swinger, K.K, Schenkel, L.B, Church, W.D, Perl, N.R, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2021-01-05 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Chembiochem, 22, 2021
|
|
7AM1
 
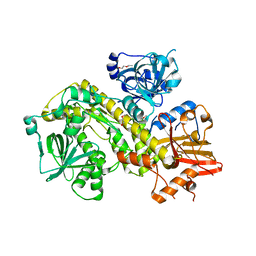 | | Structure of yeast Ssd1, a pseudonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, Protein SSD1 | | Authors: | Cook, A.G, Jayachandran, U. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Yeast Ssd1 is a non-enzymatic member of the RNase II family with an alternative RNA recognition site.
Nucleic Acids Res., 50, 2022
|
|
7KGX
 
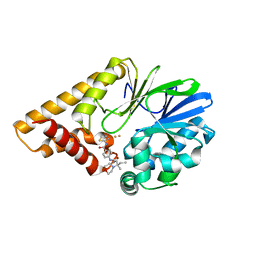 | | Structure of PQS Response Protein PqsE in Complex with 4-(3-(2-methyl-2-morpholinobutyl)ureido)-N-(thiazol-2-yl)benzamide | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-({[(2R)-2-methyl-2-(morpholin-4-yl)butyl]carbamoyl}amino)-N-(1,3-thiazol-2-yl)benzamide, FE (III) ION | | Authors: | Jeffrey, P.D, Taylor, I.R, Bassler, B.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor Mimetic Mutations in the Pseudomonas aeruginosa PqsE Enzyme Reveal a Protein-Protein Interaction with the Quorum-Sensing Receptor RhlR That Is Vital for Virulence Factor Production.
Acs Chem.Biol., 16, 2021
|
|
5VH6
 
 | | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis. | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.
To Be Published
|
|
3BEJ
 
 | | Structure of human FXR in complex with MFA-1 and co-activator peptide | | Descriptor: | (8alpha,10alpha,13alpha,17beta)-17-[(4-hydroxyphenyl)carbonyl]androsta-3,5-diene-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Soisson, S.M, Parthasarathy, G, Becker, J.W. | | Deposit date: | 2007-11-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a potent synthetic FXR agonist with an unexpected mode of binding and activation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1KJ2
 
 | | Murine Alloreactive ScFv TCR-Peptide-MHC Class I Molecule Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Allogeneic H-2Kb MHC Class I Molecule, Beta-2 microglobulin, ... | | Authors: | Reiser, J.-B, Gregoire, C, Darnault, C, Mosser, T, Guimezanes, A, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Mazza, G, Malissen, B, Housset, D. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex.
Immunity, 16, 2002
|
|
4LLG
 
 | | Crystal Structure Analysis of the E.coli holoenzyme/Gp2 complex | | Descriptor: | Bacterial RNA polymerase inhibitor, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-09 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7936 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7KXQ
 
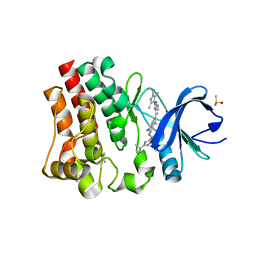 | | BTK1 SOAKED WITH COMPOUND 30 | | Descriptor: | 3-tert-butyl-N-[(5R)-2-{2-[3,5-dimethyl-1-(propan-2-yl)-1H-pyrazol-4-yl]-3H-imidazo[4,5-b]pyridin-7-yl}-6,7,8,9-tetrahydro-5H-benzo[7]annulen-5-yl]-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Isoform BTK-C of Tyrosine-protein kinase BTK | | Authors: | Viacava Follis, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
156L
 
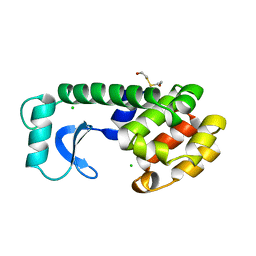 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
5AIU
 
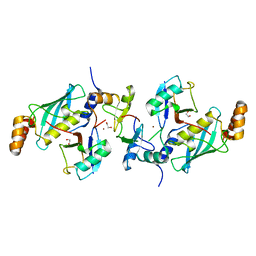 | | A complex of RNF4-RING domain, Ubc13-Ub (isopeptide crosslink) | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, ... | | Authors: | Branigan, E, Naismith, J.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
7BHW
 
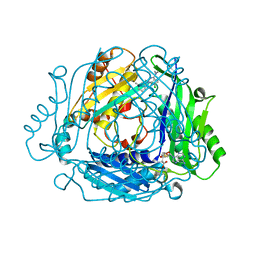 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 29) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-(3-methylphenyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
8RI3
 
 | | Crystal structure of transplatin/B-DNA adduct obtained upon 7 days of soaking | | Descriptor: | AMMONIA, CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), ... | | Authors: | Tito, G, Troisi, R, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
7BHV
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor and in vivo tool compound 28 | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-phenyl-quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
5VEG
 
 | | Structure of a Short-Chain Flavodoxin Associated with a Non-Canonical PDU Bacterial Microcompartment | | Descriptor: | CADMIUM ION, FLAVIN MONONUCLEOTIDE, Flavodoxin, ... | | Authors: | Sutter, M, Plegaria, J.S, Kerfeld, C.A. | | Deposit date: | 2017-04-04 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Functional Characterization of a Short-Chain Flavodoxin Associated with a Noncanonical 1,2-Propanediol Utilization Bacterial Microcompartment.
Biochemistry, 56, 2017
|
|
