5ZWH
 
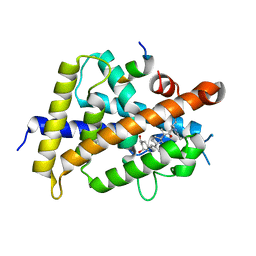 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having an ene-ynone group via conjugate addition reaction | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, (E,2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-6-en-4-yn-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, ... | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5ZXB
 
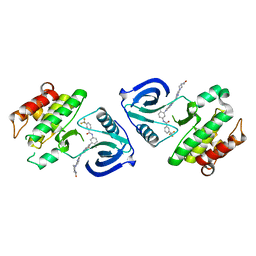 | | Crystal structure of ACK1 with compound 10d | | Descriptor: | Activated CDC42 kinase 1, N-{3-[7-{[6-(4-acetylpiperazin-1-yl)pyridin-3-yl]amino}-1-methyl-2-oxo-1,4-dihydropyrimido[4,5-d]pyrimidin-3(2H)-yl]-4-methylphenyl}-3-(trifluoromethyl)benzamide | | Authors: | Hong, E.M, Kim, H.L, Sim, T.B. | | Deposit date: | 2018-05-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | First SAR Study for Overriding NRAS Mutant Driven Acute Myeloid Leukemia.
J. Med. Chem., 61, 2018
|
|
1V3K
 
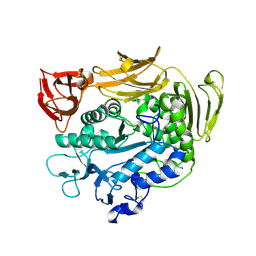 | | Crystal structure of F283Y mutant cyclodextrin glycosyltransferase | | Descriptor: | CALCIUM ION, Cyclomaltodextrin glucanotransferase | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2003-11-03 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Phe283 in enzymatic reaction of cyclodextrin glycosyltransferase from alkalophilic Bacillus sp.1011: substrate binding and arrangement of the catalytic site
PROTEIN SCI., 13, 2004
|
|
8WE7
 
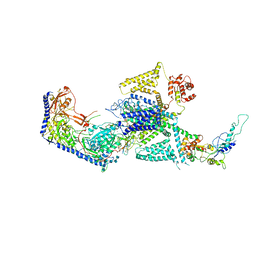 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine at 3.2 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
3E33
 
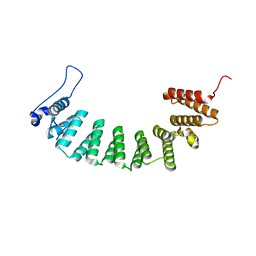 | | Protein farnesyltransferase complexed with FPP and ethylenediamine scaffold inhibitor 7 | | Descriptor: | FARNESYL DIPHOSPHATE, N-benzyl-N-(2-{(4-cyanophenyl)[(1-methyl-1H-imidazol-5-yl)methyl]amino}ethyl)-2-methylbenzenesulfonamide, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
2WKE
 
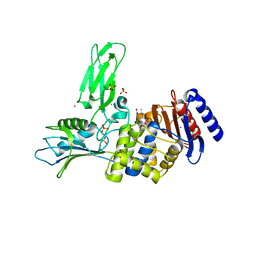 | | Crystal structure of the Actinomadura R39 DD-peptidase inhibited by 6- beta-iodopenicillanate. | | Descriptor: | (3S)-2,2-dimethyl-3,4-dihydro-2H-1,4-thiazine-3,6-dicarboxylic acid, COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, ... | | Authors: | Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2009-06-10 | | Release date: | 2009-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of the Inhibition of Class a Beta-Lactamases and Penicillin-Binding Proteins by 6-Beta-Iodopenicillanate.
J.Am.Chem.Soc., 131, 2009
|
|
4ZKV
 
 | |
7BDO
 
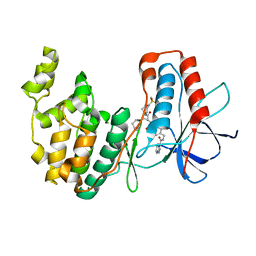 | | MAPK14 bound with SR302 | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{S})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Schroeder, M, Roehm, S, Joerger, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
5T7B
 
 | |
7BBP
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone H4, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac
To Be Published
|
|
5D2M
 
 | | Complex between human SUMO2-RANGAP1, UBC9 and ZNF451 | | Descriptor: | 1,2-ETHANEDIOL, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Cappadocia, L, Lima, C.D. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4CF3
 
 | |
3Q9S
 
 | |
5D3N
 
 | | First bromodomain of BRD4 bound to inhibitor XD40 | | Descriptor: | 4-acetyl-3-ethyl-5-methyl-N-[2-methyl-5-(methylsulfamoyl)phenyl]-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
4C1R
 
 | | Bacteroides thetaiotaomicron VPI-5482 mannosyl-6-phosphatase Bt3783 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MANNOSYL-6-PHOSPHATASE | | Authors: | Cuskin, F, Lowe, E.C, Zhu, Y, Temple, M, Thompson, A.J, Cartmell, A, Piens, K, Bracke, D, Vervecken, W, Munoz-Munoz, J.L, Suits, M.D.L, Boraston, A.B, Williams, S.J, Davies, G.J, Abbott, W.D, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
2PLT
 
 | |
4P92
 
 | | Crystal structure of dienelactone hydrolase C123S mutant at 1.65 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Carr, P.D, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization of dienelactone hydrolase in two space groups: structural changes caused by crystal packing.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4JEX
 
 | | Y21K mutant of N-acetylornithine aminotransferase complexed with L-canaline | | Descriptor: | (2S)-2-azanyl-4-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxy-butanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
7U4H
 
 | |
6AD9
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-9 | | Descriptor: | 12-mer peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, 3-[(1E)-1-{8-[(4-methyl-2-propyl-1H-benzimidazol-1-yl)methyl]dibenzo[b,e]oxepin-11(6H)-ylidene}ethyl]-1,2,4-oxadiazol-5(4H)-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Takahashi, Y, Suzuki, M, Yamamoto, K, Saito, J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Dihydrodibenzooxepine Peroxisome Proliferator-Activated Receptor (PPAR) Gamma Ligands of a Novel Binding Mode as Anticancer Agents: Effective Mimicry of Chiral Structures by Olefinic E/ Z-Isomers.
J. Med. Chem., 61, 2018
|
|
5TNK
 
 | |
6TM9
 
 | |
1V7A
 
 | | Crystal structures of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-2-HYDROXY-1-[2-(2-NAPHTHYLOXY)ETHYL]PROPYL}-1H-IMIDAZONE-4-CARBOXAMIDE, ZINC ION, adenosine deaminase | | Authors: | Kinoshita, T. | | Deposit date: | 2003-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
7FTW
 
 | |
5DBQ
 
 | | Crystal structure of insect thioredoxin at 1.95 Angstroms | | Descriptor: | Thioredoxin | | Authors: | Klinke, S, Tejedor, M.D, Cerutti, M.L, Giacometti, R, Otero, L.H, Goldbaum, F.A, Zavala, J.A, Wolosiuk, R.A, Pagano, E.A. | | Deposit date: | 2015-08-21 | | Release date: | 2016-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of insect thioredoxin at 1.95 Angstroms
To Be Published
|
|
