4NG6
 
 | | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Tsoumpra, M.K, Muniz, J.R.C, Barnett, B.L, Pilka, E, Kwaasi, A.A, Kavanagh, K.L, Evdokimov, A, Walter, R.L, Ebetino, F.H, Oppermann, U, Russell, R.G.G, Dunford, J.E. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates
TO BE PUBLISHED
|
|
6PUX
 
 | |
1RQH
 
 | | Propionibacterium shermanii transcarboxylase 5S subunit bound to pyruvic acid | | Descriptor: | COBALT (II) ION, PYRUVIC ACID, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-12-05 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
4NEX
 
 | | Structure of the N-acetyltransferase domain of X. fastidiosa NAGS/K | | Descriptor: | Acetylglutamate kinase, N-ACETYL-L-GLUTAMATE, SULFATE ION | | Authors: | Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2013-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6955 Å) | | Cite: | Structures of the N-acetyltransferase domain of Xylella fastidiosaN-acetyl-L-glutamate synthase/kinase with and without a His tag bound to N-acetyl-L-glutamate.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
5TDS
 
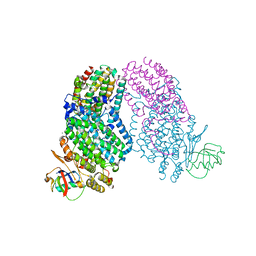 | |
5TDU
 
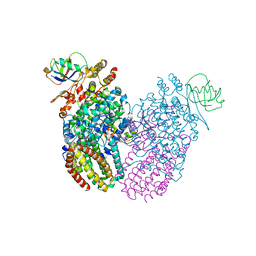 | |
4JH4
 
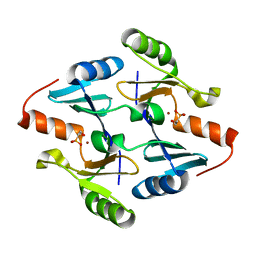 | | Crystal Structure of FosB from Bacillus cereus with Nickel and Fosfomycin | | Descriptor: | FOSFOMYCIN, Metallothiol transferase FosB, NICKEL (II) ION | | Authors: | Thompson, M.K, Harp, J, Keithly, M.E, Jagessar, K, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-03-04 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Chemical Aspects of Resistance to the Antibiotic Fosfomycin Conferred by FosB from Bacillus cereus.
Biochemistry, 52, 2013
|
|
5YXC
 
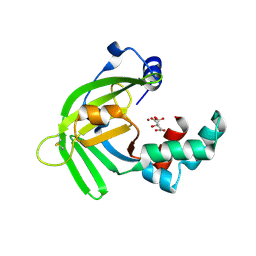 | | Crystal structure of Zinc binding protein ZinT in complex with citrate from E. coli | | Descriptor: | CITRIC ACID, Metal-binding protein ZinT, ZINC ION | | Authors: | Chen, J, Wang, L, Shang, F, Xu, Y. | | Deposit date: | 2017-12-04 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal structure of E. coli ZinT with one zinc-binding mode and complexed with citrate
Biochem. Biophys. Res. Commun., 500, 2018
|
|
4NKE
 
 | | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates | | Descriptor: | 1,2-ETHANEDIOL, 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Tsoumpra, M.K, Barnett, B.L, Muniz, J.R.C, Walter, R.L, Ebetino, F.H, von Delft, F, Russell, R.G.G, Oppermann, U, Dunford, J.E. | | Deposit date: | 2013-11-12 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates
To be Published
|
|
5V33
 
 | | R. sphaeroides photosythetic reaction center mutant - Residue L223, Ser to Trp - Room Temperature Structure Solved on X-ray Transparent Microfluidic Chip | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Schieferstein, J.M, Pawate, A.S, Sun, C, Wan, F, Broecker, J, Ernst, O.P, Gennis, R.B, Kenis, P.J.A. | | Deposit date: | 2017-03-06 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | X-ray transparent microfluidic chips for high-throughput screening and optimization of in meso membrane protein crystallization.
Biomicrofluidics, 11, 2017
|
|
1R57
 
 | | NMR Solution Structure of a GCN5-like putative N-acetyltransferase from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR31 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Acton, T.B, Ma, L, Xiao, R.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
1R62
 
 | | Crystal structure of the C-terminal Domain of the Two-Component System Transmitter Protein NRII (NtrB) | | Descriptor: | Nitrogen regulation protein NR(II) | | Authors: | Song, Y, Peisach, D, Pioszak, A.A, Xu, Z, Ninfa, A.J. | | Deposit date: | 2003-10-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the C-terminal Domain of the Two-Component System Transmitter Protein Nitrogen Regulator II (NRII; NtrB), Regulator of Nitrogen Assimilation in Escherichia coli.
Biochemistry, 43, 2004
|
|
4JRM
 
 | | Crystal structure of beta-ketoacyl-ACP synthase II (FabF) from Vibrio Cholerae (space group P212121) at 1.75 Angstrom | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, ACETATE ION, GLYCEROL | | Authors: | Hou, J, Chruszcz, M, Shabalin, I.G, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of beta-ketoacyl-ACP synthase II (FabF) from Vibrio cholerae (space group P43) at 2.2 Angstrom
To be Published
|
|
2ZW7
 
 | |
4JH3
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and Fosfomycin | | Descriptor: | FORMIC ACID, FOSFOMYCIN, GLYCEROL, ... | | Authors: | Thompson, M.K, Harp, J, Keithly, M.E, Jagessar, K, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-03-04 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Chemical Aspects of Resistance to the Antibiotic Fosfomycin Conferred by FosB from Bacillus cereus.
Biochemistry, 52, 2013
|
|
6CW2
 
 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 1 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2ZSU
 
 | |
1JU6
 
 | | Human Thymidylate Synthase Complex with dUMP and LY231514, A Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, PHOSPHATE ION, ... | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-23 | | Release date: | 2001-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
4NFK
 
 | | Crystal structure of human FPPS in complex with nickel, JDS05120, and sulfate | | Descriptor: | Farnesyl pyrophosphate synthase, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Park, J, De schutter, J.W, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
6Q03
 
 | | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine) | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine)
To Be Published
|
|
2ZW5
 
 | |
1RGN
 
 | | Structure of the reaction centre from Rhodobacter sphaeroides carotenoidless strain R-26.1 reconstituted with spheroidene | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Roszak, A.W, Frank, H.A, McKendrick, K, Mitchell, I.A, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-11-12 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Regulation of Carotenoid Binding: Gatekeeper and Locking Amino Acid Residues in Reaction Centers of Rhodobacter sphaeroides
STRUCTURE, 12, 2004
|
|
2ZCQ
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-652 | | Descriptor: | (1R)-4-(3-phenoxyphenyl)-1-phosphonobutane-1-sulfonic acid, Dehydrosqualene synthase, MAGNESIUM ION | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H, Oldfield, E. | | Deposit date: | 2007-11-11 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence.
Science, 319, 2008
|
|
4JH6
 
 | | Crystal Structure of FosB from Bacillus cereus with Manganese and Fosfomycin | | Descriptor: | FOSFOMYCIN, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Thompson, M.K, Harp, J, Keithly, M.E, Jagessar, K, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-03-04 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and Chemical Aspects of Resistance to the Antibiotic Fosfomycin Conferred by FosB from Bacillus cereus.
Biochemistry, 52, 2013
|
|
4LWY
 
 | | L(M196)H,H(M202)L Double Mutant Structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Gabdulkhakov, A.G. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Molecular Dynamic Studies of Reaction Centers Mutants from Rhodobacter sphaeroides and his mutant form L(M196)H+H(M202)L
CRYSTALLOGR REP., 59, 2014
|
|
