5F6U
 
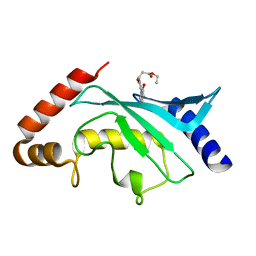 | | Crystal Structure of Ubc9 (K48A/K49A/E54A) complexed with Fragment 8 (JSS190B146) | | Descriptor: | SUMO-conjugating enzyme UBC9, ethyl 3-[4-(2-hydroxyphenyl)-3-oxidanyl-phenyl]propanoate | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, Z, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5WKB
 
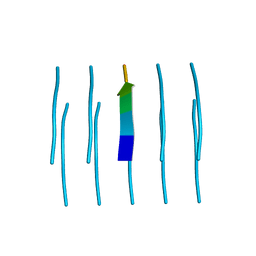 | | MicroED structure of the segment, NFGEFS, from the A315E familial variant of the low complexity domain of TDP-43, residues 312-317 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2017-07-24 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7JXB
 
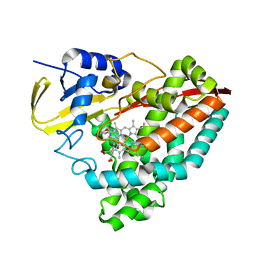 | | The crystal structure of 4-(3'-methoxyphenyl)benzoic acid-bound CYP199A4 | | Descriptor: | 3'-methoxy[1,1'-biphenyl]-4-carboxylic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Doherty, D.Z, Bell, S.G, Bruning, J. | | Deposit date: | 2020-08-27 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Enabling Aromatic Hydroxylation in a Cytochrome P450 Monooxygenase Enzyme through Protein Engineering.
Chemistry, 28, 2022
|
|
5EZ9
 
 | |
4D9S
 
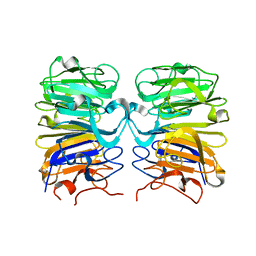 | | Crystal structure of Arabidopsis thaliana UVR8 (UV Resistance locus 8) | | Descriptor: | UVB-resistance protein UVR8 | | Authors: | Arvai, A.S, Christie, J.M, Pratt, A.J, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Plant UVR8 Photoreceptor Senses UV-B by Tryptophan-Mediated Disruption of Cross-Dimer Salt Bridges.
Science, 335, 2012
|
|
1RFX
 
 | | Crystal Structure of resisitin | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-10 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
2GMO
 
 | | NMR-structure of an independently folded C-terminal domain of influenza polymerase subunit PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Boudet, J, Tarendeau, F, Guilligay, D, Mas, P, Bougault, C.M, Cusack, S, Simorre, J.-P, Hart, D.J. | | Deposit date: | 2006-04-07 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3KF2
 
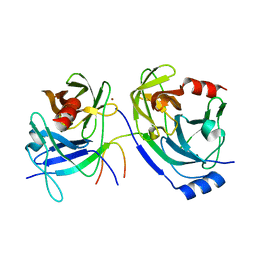 | | The HCV NS3/NS4A protease apo structure | | Descriptor: | 19-mer peptide from Genome polyprotein, Polyprotein, ZINC ION | | Authors: | Lindberg, J.D, Nystrom, S, Cummings, M.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Induced-Fit Binding of the Macrocyclic Noncovalent Inhibitor TMC435 to its HCV NS3/NS4A Protease Target
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2AGL
 
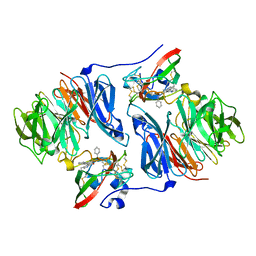 | | Crystal structure of the phenylhydrazine adduct of aromatic amine dehydrogenase from Alcaligenes faecalis | | Descriptor: | 1-PHENYLHYDRAZINE, Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
2AGY
 
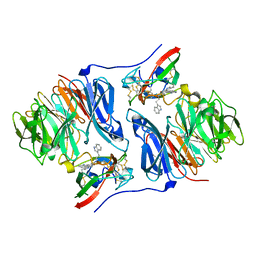 | | Crystal structure of the Schiff base intermediate in the reductive half-reaction of aromatic amine dehydrogenase (AADH) with tryptamine. Monoclinic form | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANIMINE, Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
4D8Z
 
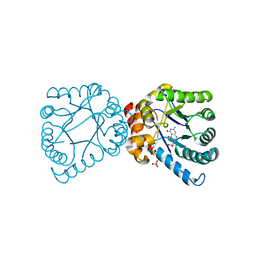 | | Crystal structure of B. anthracis DHPS with compound 24 | | Descriptor: | (3R)-3-(7-amino-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)butanoic acid, Dihydropteroate Synthase, SULFATE ION | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
2AVU
 
 | | Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription | | Descriptor: | Flagellar transcriptional activator flhC, Transcriptional activator flhD, ZINC ION | | Authors: | Wang, S, Fleming, R.T, Westbrook, E.M, Matsumura, P, McKay, D.B. | | Deposit date: | 2005-08-30 | | Release date: | 2005-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Escherichia coli FlhDC Complex, a Prokaryotic Heteromeric Regulator of Transcription.
J.Mol.Biol., 355, 2006
|
|
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
2GK6
 
 | | Structural and Functional insights into the human Upf1 helicase core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Cheng, Z, Muhlrad, D, Parker, R, Song, H. | | Deposit date: | 2006-03-31 | | Release date: | 2007-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the human Upf1 helicase core
Embo J., 26, 2007
|
|
4D7E
 
 | | An unprecedented NADPH domain conformation in Lysine Monooxygenase NbtG from Nocardia farcinica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-LYS MONOOXYGENASE | | Authors: | Binda, C, Robinson, R, Keul, N, Rodriguez, P, Robinson, H.H, Mattevi, A, Sobrado, P. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Unprecedented Nadph Domain Conformation in Lysine Monooxygenase Nbtg Provides Insights Into Uncoupling of Oxygen Consumption from Substrate Hydroxylation.
J.Biol.Chem., 290, 2015
|
|
1RRK
 
 | | Crystal Structure Analysis of the Bb segment of Factor B | | Descriptor: | COBALT (II) ION, Complement factor B, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-08 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
3N1R
 
 | | Crystal Structure of ShhN | | Descriptor: | CALCIUM ION, Sonic hedgehog protein, ZINC ION | | Authors: | Kavran, J.M, Leahy, D.J. | | Deposit date: | 2010-05-16 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of ShhN
TO BE PUBLISHED
|
|
1RS0
 
 | | Crystal Structure Analysis of the Bb segment of Factor B complexed with Di-isopropyl-phosphate (DIP) | | Descriptor: | Complement factor B, DIISOPROPYL PHOSPHONATE, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-09 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
2Z1I
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2Z1W
 
 | | tRNA guanine transglycosylase TGT E235Q mutant in complex with BDI (2-BUTYL-5,6-DIHYDRO-1H-IMIDAZO[4,5-D]PYRIDAZINE-4,7-DIONE) | | Descriptor: | 2-BUTYL-5,6-DIHYDRO-1H-IMIDAZO[4,5-D]PYRIDAZINE-4,7-DIONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Stengl, B, Heine, A, Garcia, G.A, Klebe, G, Reuter, K. | | Deposit date: | 2007-05-16 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Glutamate versus Glutamine Exchange Swaps Substrate Selectivity in tRNA-Guanine Transglycosylase: Insight into the Regulation of Substrate Selectivity by Kinetic and Crystallographic Studies
J.Mol.Biol., 374, 2007
|
|
3N1P
 
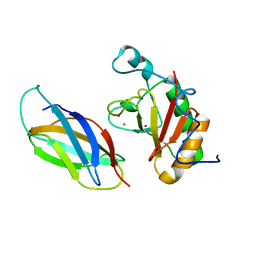 | | Crystal Structure of IhhN bound to BOCFn3 | | Descriptor: | Brother of CDO, CALCIUM ION, Indian hedgehog protein, ... | | Authors: | Kavran, J.M, Leahy, D.J. | | Deposit date: | 2010-05-16 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner.
J.Biol.Chem., 285, 2010
|
|
4GOU
 
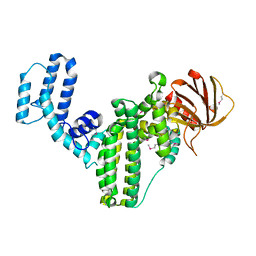 | | Crystal structure of an RGS-RhoGEF from Entamoeba histolytica | | Descriptor: | EhRGS-RhoGEF | | Authors: | Bosch, D.E, Kimple, A.J, Muller, R.E, Willard, F.S, Machius, M, Siderovski, D.P. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants of RGS-RhoGEF Signaling Critical to Entamoeba histolytica Pathogenesis.
Structure, 21, 2013
|
|
3N4Q
 
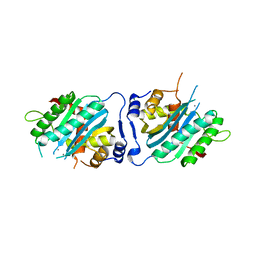 | | Human cytomegalovirus terminase nuclease domain, Mn soaked | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, TERMINASE SUBUNIT UL89 PROTEIN | | Authors: | Nadal, M, Mas, P.J, Blanco, A.G, Arnan, C, Sola, M, Hart, D.J, Coll, M. | | Deposit date: | 2010-05-22 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1RQ0
 
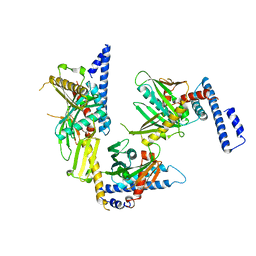 | | Crystal structure of peptide releasing factor 1 | | Descriptor: | Peptide chain release factor 1 | | Authors: | Shin, D.H, Brandsen, J, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-12-03 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome.
J.Mol.Biol., 341, 2004
|
|
