2N3Z
 
 | | Solution NMR Structure of de novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | OR446 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-15 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
2N4E
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34 | | Descriptor: | OR34 | | Authors: | Liu, G, Chan, K, Basanta, B, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-12-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34
To be Published
|
|
5K1S
 
 | | crystal structure of AibC | | Descriptor: | Oxidoreductase, zinc-binding dehydrogenase family, ZINC ION | | Authors: | Bock, T, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of AibC, a reductase involved in alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4PA8
 
 | | Crystal structure of a de novo retro-aldolase catalyzing asymmetric Michael additions, with a covalently bound product analog | | Descriptor: | (3R)-3-(4-methoxyphenyl)-5-oxohexanenitrile, GLYCEROL, SULFATE ION, ... | | Authors: | Beck, T, Garrabou Pi, X, Hilvert, D. | | Deposit date: | 2014-04-07 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Promiscuous De Novo Retro-Aldolase Catalyzes Asymmetric Michael Additions via Schiff Base Intermediates.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2N41
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34 | | Descriptor: | OR34 | | Authors: | Liu, G, Chan, K, Basanta, B, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34
To be Published
|
|
8WM0
 
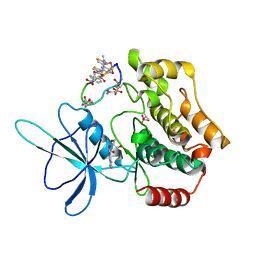 | | Crystal structure of TNIK-thiopeptide wTP3 complex | | Descriptor: | ADENOSINE, THIOPEPTIDE wTP3, TRAF2 and NCK-interacting protein kinase | | Authors: | Hamada, K, Kobayashi, S, Vinogradov, A.A, Zhang, Y, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-10-01 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides.
J.Am.Chem.Soc., 2024
|
|
5JD2
 
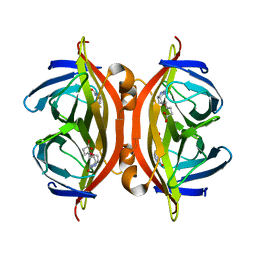 | | SFX structure of corestreptavidin-selenobiotin complex | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-selenopheno[3,4-d]imidazol-4-yl]pentanoic acid, Streptavidin | | Authors: | DeMirci, H, Hunter, M.S, Boutet, S. | | Deposit date: | 2016-04-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selenium single-wavelength anomalous diffraction de novo phasing using an X-ray-free electron laser.
Nat Commun, 7, 2016
|
|
6MNZ
 
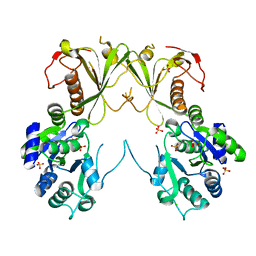 | | Crystal structure of RibBX, a two domain 3,4-dihydroxy-2-butanone 4-phosphate synthase from A. baumannii. | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Wang, J, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Multi-metal Restriction by Calprotectin Impacts De Novo Flavin Biosynthesis in Acinetobacter baumannii.
Cell Chem Biol, 26, 2019
|
|
8PK9
 
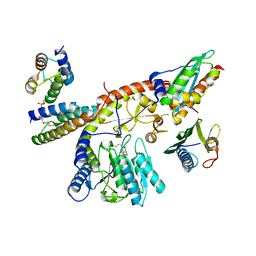 | |
8PKA
 
 | |
8PK8
 
 | |
8JMN
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-21 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[[2-[(4-chlorophenyl)methoxy]phenyl]methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-06-05 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8JSL
 
 | | The structure of EBOV L-VP35-RNA complex | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV, ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8JSM
 
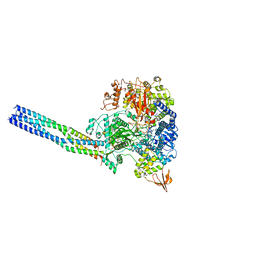 | | The structure of EBOV L-VP35-RNA complex (conformation 1) | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome., ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8JSN
 
 | | The structure of EBOV L-VP35-RNA complex (conformation 2) | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome, ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
6W3F
 
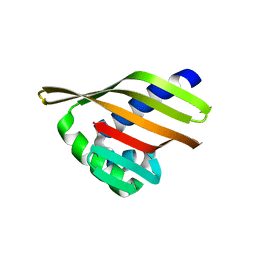 | |
6W3D
 
 | | Rd1NTF2_05 with long sheet | | Descriptor: | Rd1NTF2_05 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W3G
 
 | | Rd1NTF2_04 with long sheet | | Descriptor: | Rd1NTF2_04 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6BRR
 
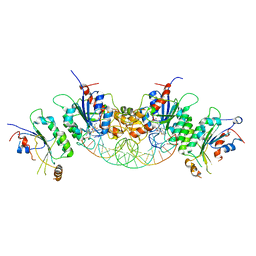 | |
6DHZ
 
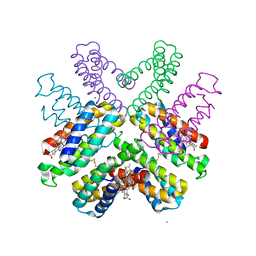 | |
6TQI
 
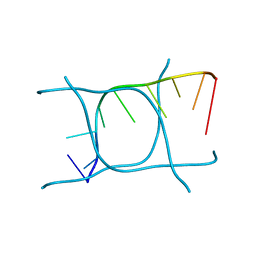 | | I-MOTIF STRUCTURE FORMED FROM THE C STRAND OF A HUMAN TELOMERE FRAGMENT | | Descriptor: | DNA (5'-*TP*AP*AP*CP*CP*CP*TP*AP*A-3') | | Authors: | Parkinson, G.N, Wagner, A, Viladoms-Claverol, J, Duman, R, El-Omari, K. | | Deposit date: | 2019-12-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6DHY
 
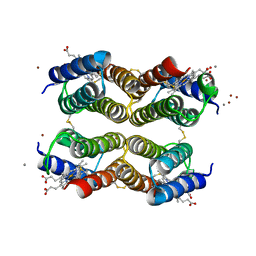 | |
7BX2
 
 | |
5K7H
 
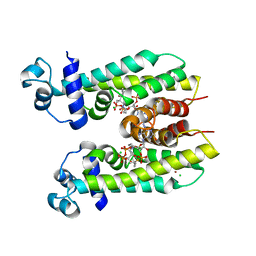 | | Crystal structure of AibR in complex with the effector molecule isovaleryl coenzyme A | | Descriptor: | CHLORIDE ION, Isovaleryl-coenzyme A, NICKEL (II) ION, ... | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
5KEZ
 
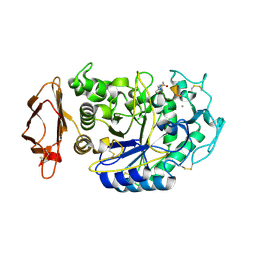 | |
