2FIF
 
 | |
2FNJ
 
 | |
2FID
 
 | |
2G45
 
 | | Co-crystal structure of znf ubp domain from the deubiquitinating enzyme isopeptidase T (isot) in complex with ubiquitin | | Descriptor: | CHLORIDE ION, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Reyes-Turcu, F.E, Horton, J.R, Mullally, J.E, Heroux, A, Cheng, X, Wilkinson, K.D. | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
2G3Q
 
 | |
2HD5
 
 | | USP2 in complex with ubiquitin | | Descriptor: | Polyubiquitin, Ubiquitin carboxyl-terminal hydrolase 2, ZINC ION | | Authors: | Renatus, M, Kroemer, M. | | Deposit date: | 2006-06-20 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2.
Structure, 14, 2006
|
|
5ZBI
 
 | |
5ZQ7
 
 | | SidE-Ubi-NAD | | Descriptor: | ADENOSINE MONOPHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SidE, ... | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ6
 
 | | SidE-Ubi-ADPr | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
6A43
 
 | | R1EN(5-225)-ubiquitin fusion | | Descriptor: | ACETIC ACID, RNA-directed DNA polymerase homolog (R1),Polyubiquitin-C | | Authors: | Maita, N. | | Deposit date: | 2018-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Determination of Ubiquitin by Fusion to a Protein That Forms a Highly Porous Crystal Lattice
J. Am. Chem. Soc., 140, 2018
|
|
5OHN
 
 | |
6R6H
 
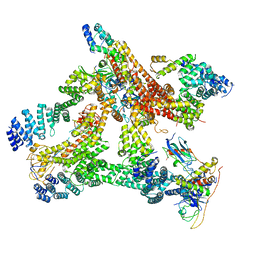 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Morris, E.P, Faull, S.V, Lau, A.M.C, Politis, A, Beuron, F, Cronin, N. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6A44
 
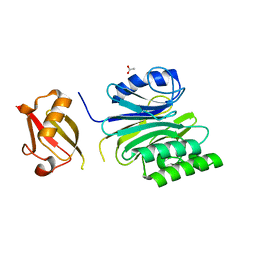 | | R1EN(5-227)-ubiquitin fusion | | Descriptor: | ACETIC ACID, RNA-directed DNA polymerase homolog (R1),Polyubiquitin-C | | Authors: | Maita, N. | | Deposit date: | 2018-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Determination of Ubiquitin by Fusion to a Protein That Forms a Highly Porous Crystal Lattice
J. Am. Chem. Soc., 140, 2018
|
|
5NVV
 
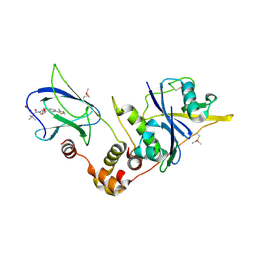 | | pVHL:EloB:EloC in complex with (2S,4R)-4-hydroxy-1-((S)-2-(2-hydroxyacetamido)-3,3-dimethylbutanoyl)-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-3,3-dimethyl-2-(2-oxidanylethanoylamino)butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
6A42
 
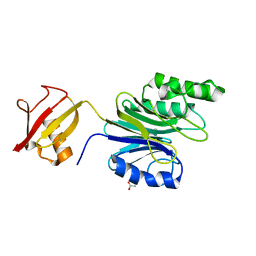 | | R1EN(5-223)-ubiquitin fusion | | Descriptor: | ACETIC ACID, RNA-directed DNA polymerase homolog (R1),Polyubiquitin-C | | Authors: | Maita, N. | | Deposit date: | 2018-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Determination of Ubiquitin by Fusion to a Protein That Forms a Highly Porous Crystal Lattice
J. Am. Chem. Soc., 140, 2018
|
|
5NW1
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(cyclobutanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 18) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-(cyclobutylcarbonylamino)-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
5NW0
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-acetamidocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 17) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-acetamidocyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
5N4W
 
 | | Crystal structure of the Cul2-Rbx1-EloBC-VHL ubiquitin ligase complex | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Cardote, T.A.F, Gadd, M.S, Ciulli, A. | | Deposit date: | 2017-02-11 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal Structure of the Cul2-Rbx1-EloBC-VHL Ubiquitin Ligase Complex.
Structure, 25, 2017
|
|
6RPQ
 
 | | Crystal structure of PhoCDC21-1 intein | | Descriptor: | Ubiquitin-like protein SMT3,1108aa long hypothetical cell division control protein | | Authors: | Beyer, H.M, Mikula, K.M, Iwai, H. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
6B7M
 
 | |
3OLM
 
 | | Structure and Function of a Ubiquitin Binding Site within the Catalytic Domain of a HECT Ubiquitin Ligase | | Descriptor: | E3 ubiquitin-protein ligase RSP5, Ubiquitin | | Authors: | Kim, H.C, Steffen, A, Chen, J, Huibregtse, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structure and function of a HECT domain ubiquitin-binding site.
Embo Rep., 12, 2011
|
|
5OHK
 
 | | Crystal structure of USP30 in covalent complex with ubiquitin propargylamide (high resolution) | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, ... | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2017-07-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mechanism and regulation of the Lys6-selective deubiquitinase USP30.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3MHS
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
5NW2
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-3,3-dimethyl-2-(oxetane-3-carboxamido)butanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 19) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-3,3-dimethyl-2-(oxetan-3-ylcarbonylamino)butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
5NVW
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(cyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 6) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-(cyclopropylcarbonylamino)-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
