3OXE
 
 | | crystal structure of glycine riboswitch, Mn2+ soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OVJ
 
 | |
3OWZ
 
 | | Crystal structure of glycine riboswitch, soaked in Iridium | | Descriptor: | Domain II of glycine riboswitch, GLYCINE, IRIDIUM HEXAMMINE ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.949 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OVL
 
 | |
3OXB
 
 | |
3OXM
 
 | | crystal structure of glycine riboswitch, Tl-Acetate soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, THALLIUM (I) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OXJ
 
 | | crystal structure of glycine riboswitch, soaked in Ba2+ | | Descriptor: | BARIUM ION, GLYCINE, MAGNESIUM ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3MOR
 
 | | Crystal structure of Cathepsin B from Trypanosoma Brucei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Cathepsin B-like cysteine protease, ... | | Authors: | Cupelli, K, Stehle, T. | | Deposit date: | 2010-04-23 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | In vivo protein crystallization opens new routes in structural biology.
Nat.Methods, 9, 2012
|
|
3LZI
 
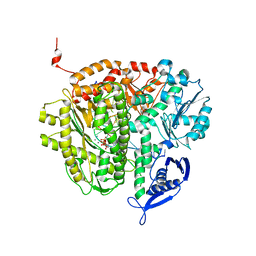 | | RB69 DNA Polymerase (Y567A) ternary complex with dATP Opposite 7,8-dihydro-8-oxoguanine | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-03-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine .
Biochemistry, 49, 2010
|
|
8JPA
 
 | |
3MZF
 
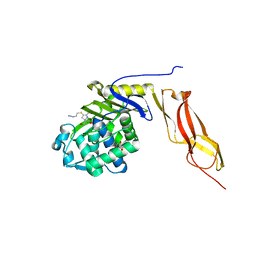 | | Structure of penicillin-binding protein 5 from E. coli: imipenem acyl-enzyme complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
3P50
 
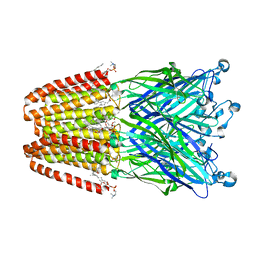 | | Structure of propofol bound to a pentameric ligand-gated ion channel, GLIC | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nury, H, Van Renterghem, C, Weng, Y, Tran, A, Baaden, M, Dufresne, V, Changeux, J.P, Sonner, J.M, Delarue, M, Corringer, P.J. | | Deposit date: | 2010-10-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel
Nature, 469, 2011
|
|
3NAD
 
 | | Crystal Structure of Phenolic Acid Decarboxylase from Bacillus pumilus UI-670 | | Descriptor: | Ferulate decarboxylase, SULFATE ION | | Authors: | Matte, A, Grosse, S, Bergeron, H, Abokitse, K, Lau, P.C.K. | | Deposit date: | 2010-06-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural analysis of Bacillus pumilus phenolic acid decarboxylase, a lipocalin-fold enzyme.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3NCI
 
 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite dG at 1.8 angstrom resolution | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Blaha, G, Steitz, T.A, Konigsberg, W.H, Wang, J. | | Deposit date: | 2010-06-04 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Insights into base selectivity from the 1.8 A resolution structure of an RB69 DNA polymerase ternary complex.
Biochemistry, 50, 2011
|
|
5FH1
 
 | | The structure of rat cytosolic PEPCK variant E89D in complex with GTP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Johnson, T.A, Holyoak, T. | | Deposit date: | 2015-12-21 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Utilization of Substrate Intrinsic Binding Energy for Conformational Change and Catalytic Function in Phosphoenolpyruvate Carboxykinase.
Biochemistry, 55, 2016
|
|
3N0I
 
 | | Crystal Structure of Ad37 fiber knob in complex with GD1a oligosaccharide | | Descriptor: | Fiber, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ZINC ION | | Authors: | Nilsson, E.C, Storm, R.J, Bauer, J, Johansson, S.M.C, Lookene, A, Angstroem, J, Hedenstroem, M, Fraengsmyr, L, Rinaldi, S, Willison, H, Domelloef, F.P, Stehle, T, Arnberg, N. | | Deposit date: | 2010-05-14 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis.
NAT.MED. (N.Y.), 17, 2011
|
|
5FH2
 
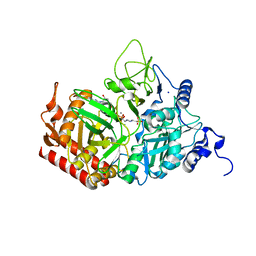 | | The structure of rat cytosolic PEPCK variant E89Q in complex with GTP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Johnson, T.A, Holyoak, T. | | Deposit date: | 2015-12-21 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Utilization of Substrate Intrinsic Binding Energy for Conformational Change and Catalytic Function in Phosphoenolpyruvate Carboxykinase.
Biochemistry, 55, 2016
|
|
3P4W
 
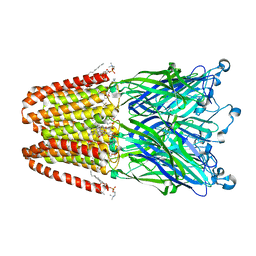 | | Structure of desflurane bound to a pentameric ligand-gated ion channel, GLIC | | Descriptor: | (2S)-2-(difluoromethoxy)-1,1,1,2-tetrafluoroethane, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nury, H, Van Renterghem, C, Weng, Y, Tran, A, Baaden, M, Dufresne, V, Changeux, J.P, Sonner, J.M, Delarue, M, Corringer, P.J. | | Deposit date: | 2010-10-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel
Nature, 469, 2011
|
|
6WJ1
 
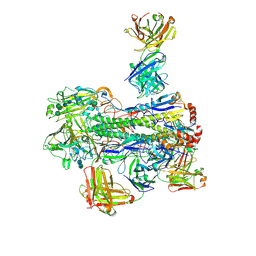 | | Crystal structure of Fab 54-4H03 bound to H1 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 54-4H03 heavy chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
3OX0
 
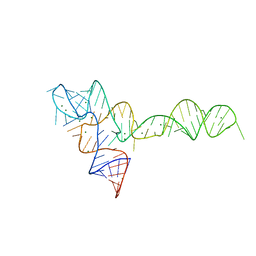 | |
5FH3
 
 | |
6WJ0
 
 | | Crystal structure of Fab 54-4H03 | | Descriptor: | Fab 54-4H03 heavy chain, Fab 54-4H03 light chain, GLYCEROL | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
3MEQ
 
 | | Crystal structure of alcohol dehydrogenase from Brucella melitensis | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase, ... | | Authors: | Arakaki, T.L, Staker, B.L, Gardberg, A, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Brucella melitensis
To be Published
|
|
3MHH
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
3LZJ
 
 | | RB69 DNA Polymerase (Y567A) ternary complex with dCTP Opposite 7,8-Dihydro-8-oxoguanine | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-03-01 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine .
Biochemistry, 49, 2010
|
|
