7SXM
 
 | |
3NUH
 
 | |
3O5T
 
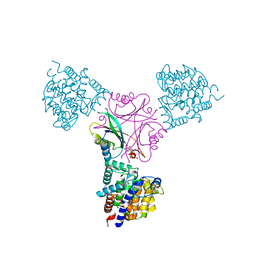 | | Structure of DraG-GlnZ complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dinitrogenase reductase activacting glicohydrolase, MAGNESIUM ION, ... | | Authors: | Rajendran, C, Li, X.-D, Winkler, F.K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of the GlnZ-DraG complex reveals a different form of PII-target interaction
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6UXA
 
 | |
5FH0
 
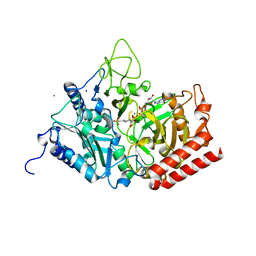 | | The structure of rat cytosolic PEPCK variant E89A complex with GTP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Johnson, T.A, Holyoak, T. | | Deposit date: | 2015-12-21 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Utilization of Substrate Intrinsic Binding Energy for Conformational Change and Catalytic Function in Phosphoenolpyruvate Carboxykinase.
Biochemistry, 55, 2016
|
|
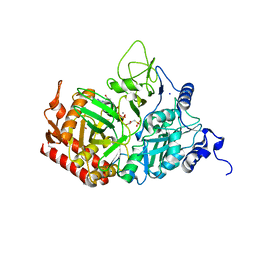
5FH4
 
 | |
3O55
 
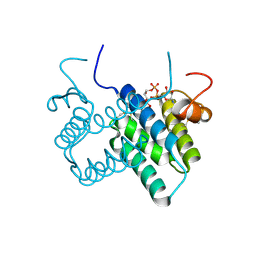 | | Crystal structure of human FAD-linked augmenter of liver regeneration (ALR) | | Descriptor: | Augmenter of liver regeneration, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Banci, L, Bertini, I, Calderone, V, Cefaro, C, Ciofi-Baffoni, S, Gallo, A, Kallergi, E, Lionaki, E, Pozidis, C, Tokatlidis, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NVE
 
 | |
3OAP
 
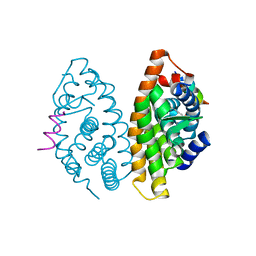 | | Crystal structure of human Retinoid X Receptor alpha-ligand binding domain complex with 9-cis retinoic acid and the coactivator peptide GRIP-1 | | Descriptor: | (9cis)-retinoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, Energetics and Dynamics of Binding Coactivator Peptide to Human Retinoid X Receptor Alpha Ligand Binding Domain Complex with 9-cis-Retinoic Acid.
Biochemistry, 50, 2011
|
|
3NVF
 
 | |
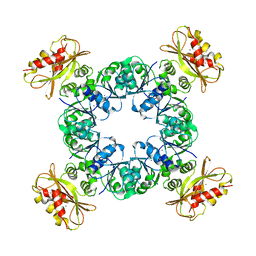
6UWN
 
 | |
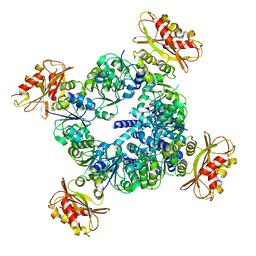
6UX7
 
 | |
3NVG
 
 | |
6UX4
 
 | |
3OGU
 
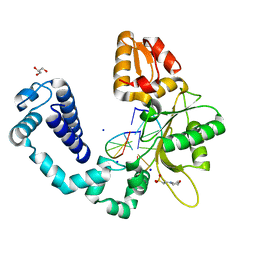 | | DNA Polymerase beta mutant 5P20 complexed with 6bp of DNA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*CP*AP*TP*CP*TP*G)-3', 5'-D(P*CP*AP*GP*AP*TP*G)-3', ... | | Authors: | Marx, A, Diederichs, K, Bergen, K. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Human DNA polymerase beta mutations allowing efficient abasic site bypass.
J.Biol.Chem., 286, 2011
|
|
3OG9
 
 | | Structure of YahD with Malic acid | | Descriptor: | D-MALATE, protein yahD a copper inducible hydrolase | | Authors: | Martinez Font, J, Mancini, S, Tauberger, E, Moniot, S. | | Deposit date: | 2010-08-16 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Regulation and structure of YahD, a copper inducible alpha/beta hydrolase of Lactococcus lactis IL1403
Fems Microbiol.Lett., 314, 2011
|
|
3OGI
 
 | | Crystal structure of the Mycobacterium tuberculosis H37Rv EsxOP complex (Rv2346c-Rv2347c) | | Descriptor: | Putative ESAT-6-like protein 6, Putative ESAT-6-like protein 7 | | Authors: | Arbing, M.A, Chan, S, Zhou, T.T, Ahn, C, Harris, L, Kuo, E, Sawaya, M.R, Cascio, D, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-08-16 | | Release date: | 2010-08-25 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
3OWI
 
 | |
3OXD
 
 | |
3OTO
 
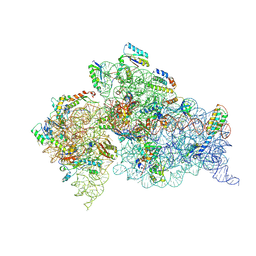 | | Crystal Structure of the 30S ribosomal subunit from a KsgA mutant of Thermus thermophilus (HB8) | | Descriptor: | 16S rRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Demirci, H, Murphy IV, F, Belardinelli, R, Kelley, A.C, Ramakrishnan, V, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2010-09-13 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function.
Rna, 16, 2010
|
|
3P0K
 
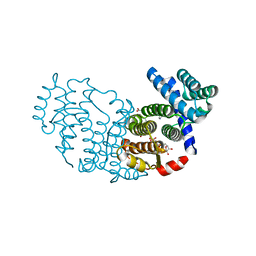 | | Structure of Baculovirus Sulfhydryl Oxidase Ac92 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Hakim, M, Fass, D. | | Deposit date: | 2010-09-29 | | Release date: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of a baculovirus sulfhydryl oxidase, a highly divergent member of the erv flavoenzyme family.
J.Virol., 85, 2011
|
|
7N71
 
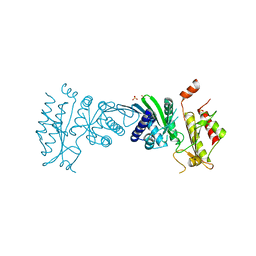 | | Crystal Structure of PI5P4KIIAlpha | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3P5S
 
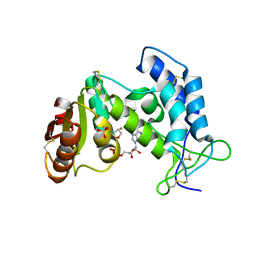 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD38 molecule, SULFATE ION, ... | | Authors: | Egea, P.F, Muller-Stauffler, H, Kohn, I, Cakou-Kefir, C, Stroud, R.M, Kellenberburger, E, Schuber, F. | | Deposit date: | 2010-10-10 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3OW9
 
 | |
3OWW
 
 | |
