5DKD
 
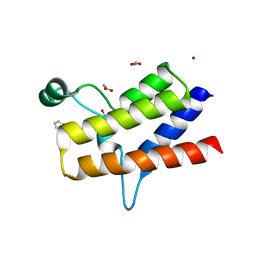 | | Crystal structure of the bromodomain of human BRG1 (SMARCA4) in complex with PFI-3 chemical probe | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-[(1R,4R)-5-(pyridin-2-yl)-2,5-diazabicyclo[2.2.1]hept-2-yl]prop-2-en-1-one, 1,2-ETHANEDIOL, Transcription activator BRG1, ... | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Fedorov, O, Savitsky, P, Nunez-Alonso, G, Newman, J.A, Filippakopoulos, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the bromodomain of human BRG1 (SMARCA4) in complex with PFI-3 chemical probe
To Be Published
|
|
5BQ5
 
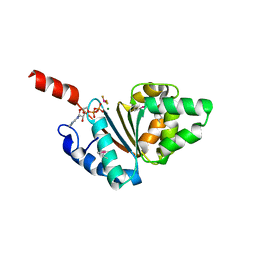 | | Crystal structure of the IstB AAA+ domain bound to ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Insertion sequence IS5376 putative ATP-binding protein, ... | | Authors: | Arias-Palomo, E, Berger, J.M. | | Deposit date: | 2015-05-28 | | Release date: | 2015-09-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Atypical AAA+ ATPase Assembly Controls Efficient Transposition through DNA Remodeling and Transposase Recruitment.
Cell, 162, 2015
|
|
4Z0X
 
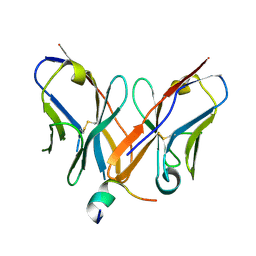 | |
4ZB6
 
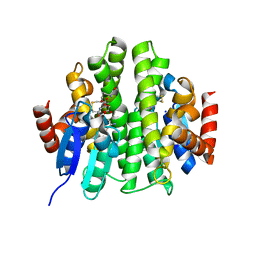 | |
5DL7
 
 | |
5DKC
 
 | | Crystal structure of the bromodomain of human BRM (SMARCA2) in complex with PFI-3 chemical probe | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-[(1R,4R)-5-(pyridin-2-yl)-2,5-diazabicyclo[2.2.1]hept-2-yl]prop-2-en-1-one, Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Fedorov, O, Savitsky, P, Nunez-Alonso, G, Fonseca, M, Krojer, T, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the bromodomain of human BRM (SMARCA2) in complex with PFI-3 chemical probe
To Be Published
|
|
4Z81
 
 | |
5E78
 
 | | Crystal structure of P450 BM3 heme domain variant complexed with Co(III)Sep | | Descriptor: | 1,3,6,8,10,13,16,19-octaazabicyclo[6.6.6]icosane, Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, ... | | Authors: | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | Deposit date: | 2015-10-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into a cobalt (III) sepulchrate based alternative cofactor system of P450 BM3 monooxygenase.
Biochim. Biophys. Acta, 1866, 2018
|
|
4ZB8
 
 | |
4ZBB
 
 | |
4ZNE
 
 | | IgG1 Fc-FcgammaRI ecd complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Oganesyan, V.Y, Dall'Acqua, W.F. | | Deposit date: | 2015-05-04 | | Release date: | 2015-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into the interaction of human IgG1 with Fc gamma RI: no direct role of glycans in binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZB9
 
 | |
5BTQ
 
 | | Crystal structure of human heme oxygenase 1 H25R with biliverdin bound | | Descriptor: | BILIVERDINE IX ALPHA, Heme oxygenase 1, SULFATE ION | | Authors: | Caaveiro, J.M.M, Morante, K, Sigala, P, Tsumoto, K. | | Deposit date: | 2015-06-03 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | In-Cell Enzymology To Probe His-Heme Ligation in Heme Oxygenase Catalysis
Biochemistry, 55, 2016
|
|
5B2Q
 
 | | Crystal structure of Francisella novicida Cas9 RHA in complex with sgRNA and target DNA (TGG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
5EDF
 
 | | Crystal structure of the selenomethionine-substituted iron-regulated protein FrpD from Neisseria meningitidis | | Descriptor: | AZIDE ION, FrpC operon protein, HEXAETHYLENE GLYCOL, ... | | Authors: | Sviridova, E, Bumba, L, Rezacova, P, Sebo, P, Kuta Smatanova, I. | | Deposit date: | 2015-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep, 7, 2017
|
|
5DZL
 
 | | Crystal structure of the protein human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Huang, Y.H, Russell, A, Gandhi, A.K, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
5E5X
 
 | |
5E2X
 
 | | The crystal structure of the C-terminal domain of Ebola (Tai Forest) nucleoprotein | | Descriptor: | NONAETHYLENE GLYCOL, NP | | Authors: | Baker, L.E, Handing, K.B, Derewenda, U, Utepbergenov, D, Derewenda, Z.S. | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular architecture of the nucleoprotein C-terminal domain from the Ebola and Marburg viruses.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5C0G
 
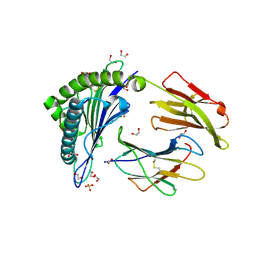 | | HLA-A02 carrying YLGGPDFPTI | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
5DL6
 
 | |
5DRZ
 
 | | Crystal structure of anti-HIV-1 antibody F240 Fab in complex with gp41 peptide | | Descriptor: | Envelope glycoprotein gp160, HIV Antibody F240 Heavy Chain, HIV Antibody F240 Light Chain, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240.
Sci Rep, 6, 2016
|
|
3MFG
 
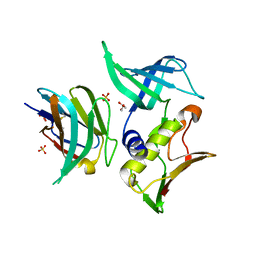 | |
3M66
 
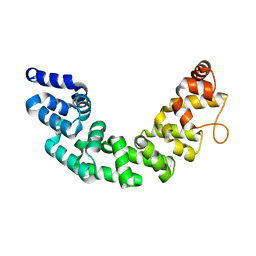 | | Crystal structure of human Mitochondrial Transcription Termination Factor 3 | | Descriptor: | mTERF domain-containing protein 1, mitochondrial | | Authors: | Spahr, H, Samuelsson, T, Hallberg, B.M, Gustafsson, C.M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain.
Biochem.Biophys.Res.Commun., 397, 2010
|
|
3MZD
 
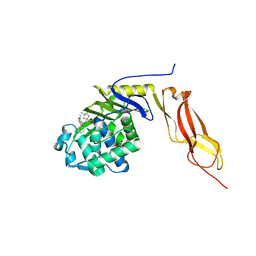 | | Structure of penicillin-binding protein 5 from E. coli: cloxacillin acyl-enzyme complex | | Descriptor: | (2R,4S)-2-[(1S)-1-({[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}amino)-2-oxoethyl]-5,5-dimethyl-1,3-thiazolid ine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
3MZE
 
 | | Structure of penicillin-binding protein 5 from E.coli: cefoxitin acyl-enzyme complex | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
