1EFV
 
 | |
1EFW
 
 | | Crystal structure of aspartyl-tRNA synthetase from Thermus thermophilus complexed to tRNAasp from Escherichia coli | | Descriptor: | ASPARTYL-TRNA, ASPARTYL-TRNA SYNTHETASE | | Authors: | Briand, C, Poterszman, A, Eiler, S, Webster, G, Thierry, J.-C, Moras, D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | An intermediate step in the recognition of tRNA(Asp) by aspartyl-tRNA synthetase.
J.Mol.Biol., 299, 2000
|
|
1EFX
 
 | | STRUCTURE OF A COMPLEX BETWEEN THE HUMAN NATURAL KILLER CELL RECEPTOR KIR2DL2 AND A CLASS I MHC LIGAND HLA-CW3 | | Descriptor: | BETA-2-MICROGLOBULIN, HLA-CW3 (HEAVY CHAIN), NATURAL KILLER CELL RECEPTOR KIR2DL2, ... | | Authors: | Boyington, J.C, Motyka, S.A, Schuck, P, Brooks, A.G, Sun, P.D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand.
Nature, 405, 2000
|
|
1EFY
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC FRAGMENT OF POLY (ADP-RIBOSE) POLYMERASE COMPLEXED WITH A BENZIMIDAZOLE INHIBITOR | | Descriptor: | 2-(3'-METHOXYPHENYL) BENZIMIDAZOLE-4-CARBOXAMIDE, POLY (ADP-RIBOSE) POLYMERASE | | Authors: | White, A.W, Almassy, R, Calvert, A.H, Curtin, N.J, Griffin, R.J, Hostomsky, Z, Maegley, K, Newell, D.R, Srinivasan, S, Golding, B.T. | | Deposit date: | 2000-02-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance-modifying agents. 9. Synthesis and biological properties of benzimidazole inhibitors of the DNA repair enzyme poly(ADP-ribose) polymerase.
J.Med.Chem., 43, 2000
|
|
1EFZ
 
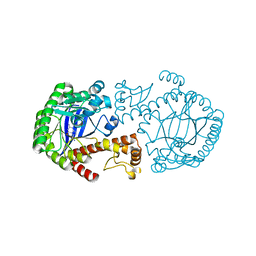 | | MUTAGENESIS AND CRYSTALLOGRAPHIC STUDIES OF ZYMOMONAS MOBILIS TRNA-GUANINE TRANSGLYCOSYLASE TO ELUCIDATE THE ROLE OF SERINE 103 FOR ENZYMATIC ACTIVITY | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Gradler, U, Ficner, R, Garcia, G.A, Stubbs, M.T, Klebe, G, Reuter, K. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase to elucidate the role of serine 103 for enzymatic activity.
FEBS Lett., 454, 1999
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
1EG1
 
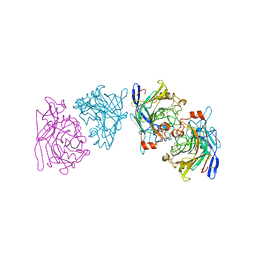 | | ENDOGLUCANASE I FROM TRICHODERMA REESEI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Kleywegt, G.J, Zou, J.-Y, Jones, T.A. | | Deposit date: | 1996-11-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 A resolution, and a comparison with related enzymes.
J.Mol.Biol., 272, 1997
|
|
1EG2
 
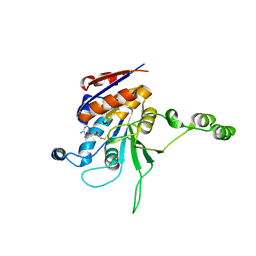 | | CRYSTAL STRUCTURE OF RHODOBACTER SPHEROIDES (N6 ADENOSINE) METHYLTRANSFERASE (M.RSRI) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MODIFICATION METHYLASE RSRI | | Authors: | Scavetta, R.D, Thomas, C.B, Walsh, M.A, Szegedi, S, Joachimiak, A, Gumport, R.I, Churchill, M.E.A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-02-11 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases.
Nucleic Acids Res., 28, 2000
|
|
1EG3
 
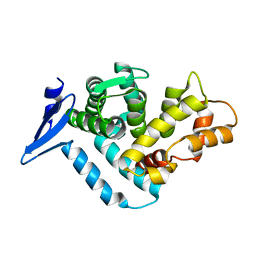 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | Descriptor: | DYSTROPHIN | | Authors: | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
1EG4
 
 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | Descriptor: | BETA-DYSTROGLYCAN, DYSTROPHIN | | Authors: | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
1EG5
 
 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Kaiser, J.T, Clausen, T, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-02-13 | | Release date: | 2000-04-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
1EG6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF D(CG(5-BRU)ACG) COMPLEXES TO A PHENAZINE | | Descriptor: | 5'-D(*CP*GP*(BRO)UP*AP*CP*G)-3', 9-BROMO-PHENAZINE-1-CARBOXYLIC ACID (2-DIMETHYLAMINO-ETHYL)-AMIDE, BROMIDE ION, ... | | Authors: | Cardin, C.J, Denny, W.A, Hobbs, J.R, Thorpe, J.H. | | Deposit date: | 2000-02-14 | | Release date: | 2001-01-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Guanine specific binding at a DNA junction formed by d[CG(5-BrU)ACG](2) with a topoisomerase poison in the presence of Co(2+) ions.
Biochemistry, 39, 2000
|
|
1EG7
 
 | | THE CRYSTAL STRUCTURE OF FORMYLTETRAHYDROFOLATE SYNTHETASE FROM MOORELLA THERMOACETICA | | Descriptor: | FORMYLTETRAHYDROFOLATE SYNTHETASE, SULFATE ION | | Authors: | Radfar, R, Shin, R, Sheldrick, G.M, Minor, W, Lovell, C.R, Odom, J.D, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-02-14 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of N(10)-formyltetrahydrofolate synthetase from Moorella thermoacetica.
Biochemistry, 39, 2000
|
|
1EG9
 
 | | NAPHTHALENE 1,2-DIOXYGENASE WITH INDOLE BOUND IN THE ACTIVE SITE. | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, INDOLE, ... | | Authors: | Carredano, E, Karlsson, A, Kauppi, B, Choudhury, D, Parales, R.E, Parales, J.V, Lee, K, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2000-02-15 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding.
J.Mol.Biol., 296, 2000
|
|
1EGA
 
 | | CRYSTAL STRUCTURE OF A WIDELY CONSERVED GTPASE ERA | | Descriptor: | PROTEIN (GTP-BINDING PROTEIN ERA), SULFATE ION | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1998-12-01 | | Release date: | 1999-07-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1EGC
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE COMPLEXED WITH OCTANOYL-COA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE, OCTANOYL-COENZYME A | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EGD
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EGE
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EGF
 
 | |
1EGG
 
 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
1EGH
 
 | |
1EGI
 
 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
1EGJ
 
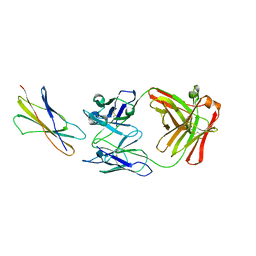 | | DOMAIN 4 OF THE BETA COMMON CHAIN IN COMPLEX WITH AN ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY (HEAVY CHAIN), ANTIBODY (LIGHT CHAIN), ... | | Authors: | Rossjohn, J, McKinstry, W.J, Woodcock, J.M, McClure, B.J, Hercus, T.R, Parker, M.W, Lopez, A.F, Bagley, C.J. | | Deposit date: | 2000-02-15 | | Release date: | 2001-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the activation domain of the GM-CSF/IL-3/IL-5 receptor common beta-chain bound to an antagonist.
Blood, 95, 2000
|
|
1EGK
 
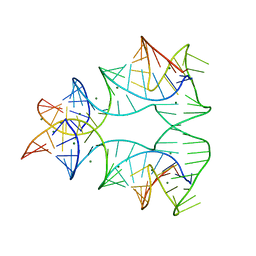 | | CRYSTAL STRUCTURE OF A NUCLEIC ACID FOUR-WAY JUNCTION | | Descriptor: | 10-23 DNA ENZYME, MAGNESIUM ION, RNA (5'-R(*AP*GP*GP*AP*GP*AP*GP*AP*GP*AP*UP*GP*GP*GP*UP*GP*CP*GP*AP*G)-3') | | Authors: | Nowakowski, J, Shim, P.J, Stout, C.D, Joyce, G.F. | | Deposit date: | 2000-02-15 | | Release date: | 2000-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Alternative conformations of a nucleic acid four-way junction.
J.Mol.Biol., 300, 2000
|
|
1EGL
 
 | |
