8I1H
 
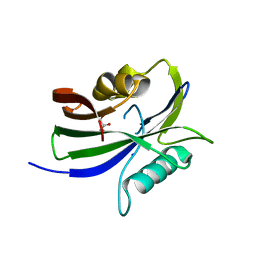 | |
6Z6N
 
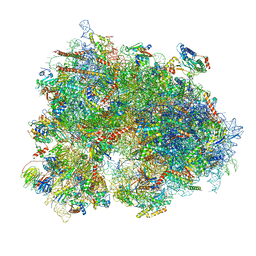 | | Cryo-EM structure of human EBP1-80S ribosomes (focus on EBP1) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6KYL
 
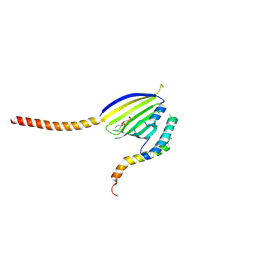 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 in Complex with (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2019-09-19 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1.
Commun Biol, 3, 2020
|
|
5M7I
 
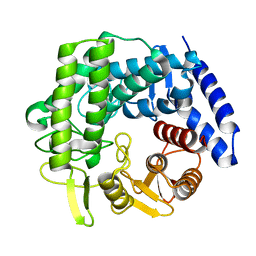 | | Crystal structure of GH125 1,6-alpha-mannosidase mutant from Clostridium perfringens in complex with 1,6-alpha-mannobiose | | Descriptor: | alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose, exo-alpha-1,6-mannosidase | | Authors: | Males, A, Alonso-Gil, S, Fernandes, P, Williams, S.J, Rovira, C, Davies, G.J. | | Deposit date: | 2016-10-27 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational Design of Experiment Unveils the Conformational Reaction Coordinate of GH125 alpha-Mannosidases.
J. Am. Chem. Soc., 139, 2017
|
|
7YWK
 
 | | Crystal structure of an engineered TycA variant, TycApPLA, in complex with AMP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Mittl, P, Camus, A, Truong, G, Markert, G, Hilvert, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Reprogramming Nonribosomal Peptide Synthetases for Site-Specific Insertion of alpha-Hydroxy Acids.
J.Am.Chem.Soc., 144, 2022
|
|
5YFZ
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and S-GS626 | | Descriptor: | 2-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-fluoranyl-phenyl]piperidin-4-yl]ethanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
4A5M
 
 | | Redox regulator HypR in its oxidized form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UNCHARACTERIZED HTH-TYPE TRANSCRIPTIONAL REGULATOR YYBR | | Authors: | Palm, G.J, Waack, P, Read, R.J, Hinrichs, W. | | Deposit date: | 2011-10-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights Into the Redox-Switch Mechanism of the Marr/Duf24-Type Regulator Hypr
Nucleic Acids Res., 40, 2012
|
|
4POQ
 
 | | Structure of unliganded VP1 pentamer of Human Polyomavirus 9 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ISOPROPYL ALCOHOL, ... | | Authors: | Khan, Z.M, Stehle, T. | | Deposit date: | 2014-02-26 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and glycan microarray analysis of human polyomavirus 9 VP1 identifies N-glycolyl neuraminic acid as a receptor candidate.
J.Virol., 88, 2014
|
|
9MMI
 
 | | Myo-inositol-1(or 4)-monophosphatase that can perform essential dephosphorylation step to facilitate riboflavin biosynthesis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Fructose-1,6-bisphosphatase/inositol-1-monophosphatase, MAGNESIUM ION, ... | | Authors: | Hoffpauir, Z.A, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2024-12-20 | | Release date: | 2025-04-09 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of the phosphatase essential for riboflavin biosynthesis in Aquifex aeolicus.
J.Biol.Chem., 301, 2025
|
|
5E96
 
 | | Crystal structure of aminoglycoside 6'-acetyltransferase type Ii | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside 6'-acetyltransferase, PHOSPHATE ION | | Authors: | Berghuis, A.M, Burk, D.L, Baettig, O.M, Shi, K. | | Deposit date: | 2015-10-14 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive characterization of ligand-induced plasticity changes in a dimeric enzyme.
Febs J., 283, 2016
|
|
7GAU
 
 | | PanDDA analysis group deposition of ground-state model of MAP1LC3B | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-11 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
1MB1
 
 | | MBP1 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | MLU1-BOX BINDING PROTEIN | | Authors: | Taylor, I.A, Smerdon, S.J. | | Deposit date: | 1997-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray structure of the DNA-binding domain from the Saccharomyces cerevisiae cell-cycle transcription factor Mbp1 at 2.1 A resolution.
J.Mol.Biol., 272, 1997
|
|
7LMT
 
 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a
To Be Published
|
|
6Q2Y
 
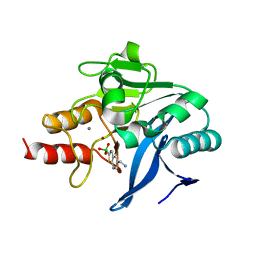 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
5DMP
 
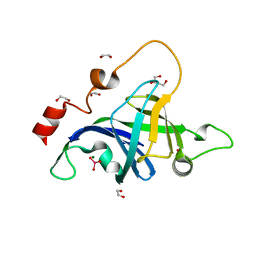 | | Structure of the Archaeal NHEJ Phosphoesterase from Methanocella paludicola. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein, ... | | Authors: | Brissett, N.C, Bartlett, E.J, Doherty, A.J. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Molecular basis for DNA strand displacement by NHEJ repair polymerases.
Nucleic Acids Res., 44, 2016
|
|
5JBS
 
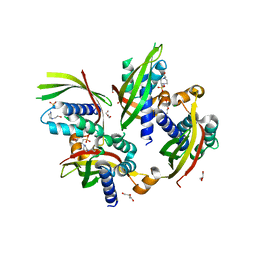 | | Conformational changes during monomer-to-dimer transition of Brucella suis VirB8 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arya, T, Sharifahmadian, M, Sygusch, J, Baron, B. | | Deposit date: | 2016-04-13 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NMR analyses, X-ray crystallography and small-molecule probing reveal conformational shifts during monomer-to-dimer transition of Brucella suis VirB8
To Be Published
|
|
3F9M
 
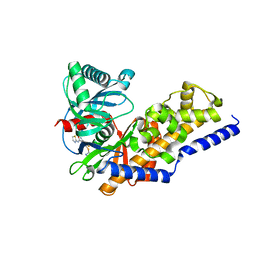 | | Human pancreatic glucokinase in complex with glucose and activator showing a mobile flap | | Descriptor: | 2-AMINO-4-FLUORO-5-[(1-METHYL-1H-IMIDAZOL-2-YL)SULFANYL]-N-(1,3-THIAZOL-2-YL)BENZAMIDE, Glucokinase, alpha-D-glucopyranose | | Authors: | Petit, P, Gluais, L, Lagarde, A, Vuillard, L, Boutin, J.A, Ferry, G. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The active conformation of human glucokinase is not altered by allosteric activators
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6ZP7
 
 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
3FA3
 
 | | Crystal structure of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member, trigonal crystal form | | Descriptor: | 2,2-difluoro-3,3-dihydroxybutanedioic acid, 2,3-dimethylmalate lyase, GLYCEROL, ... | | Authors: | Narayanan, B.C, Herzberg, O. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member.
J.Mol.Biol., 386, 2009
|
|
5FXM
 
 | |
6ZP5
 
 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
3IZX
 
 | | 3.1 Angstrom cryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Capsid protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Yu, X, Ge, P, Jiang, J, Atanasov, I, Zhou, Z.H. | | Deposit date: | 2011-01-15 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic Model of CPV Reveals the Mechanism Used by This Single-Shelled Virus to Economically Carry Out Functions Conserved in Multishelled Reoviruses.
Structure, 19, 2011
|
|
8HUM
 
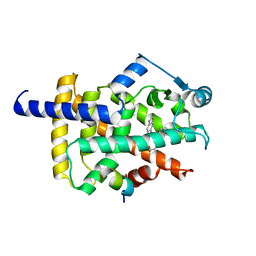 | | X-ray structure of human PPAR gamma ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
5FMJ
 
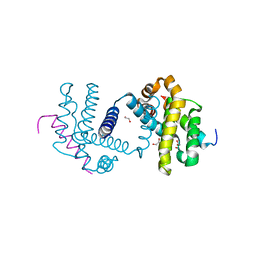 | | Bcl-xL with mouse Bak BH3 Q75L complex | | Descriptor: | 1,2-ETHANEDIOL, BAK1 PROTEIN, BCL-2-LIKE PROTEIN 1 | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Physiological Restraint of Bak by Bcl-Xl is Essential for Cell Survival.
Genes Dev., 30, 2016
|
|
6B1V
 
 | | Crystal structure of Ps i-CgsB C78S in complex with i-neocarratetraose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
