5T85
 
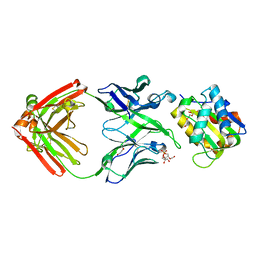 | |
7U28
 
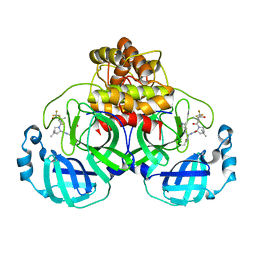 | | Structure of SARS-CoV-2 Mpro Lambda (G15S) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Greasley, S.E, Ferre, R.A, Plotnikova, O, Liu, W, Stewart, A.E. | | Deposit date: | 2022-02-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
7U29
 
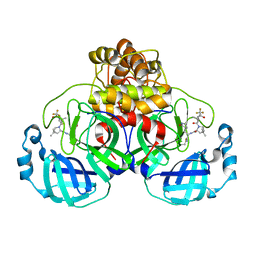 | | Structure of SARS-CoV-2 Mpro mutant (K90R) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2022-02-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
9D3G
 
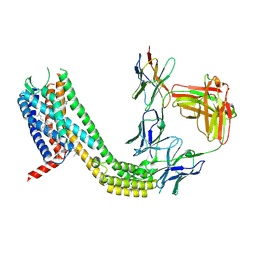 | | Cryo-EM structure of CCR6 bound by SQA1 and OXM1 | | Descriptor: | 1-(4-chlorophenyl)-N-{[(2R)-4-(2,3-dihydro-1H-inden-2-yl)-5-oxomorpholin-2-yl]methyl}cyclopropane-1-carboxamide, 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, CCR6, ... | | Authors: | Wasilko, D.J, Wu, H. | | Deposit date: | 2024-08-10 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for CCR6 modulation by allosteric antagonists.
Nat Commun, 15, 2024
|
|
5X8G
 
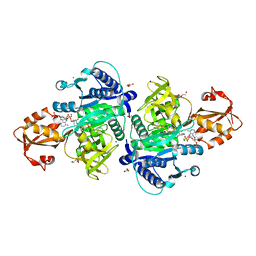 | |
1N1Z
 
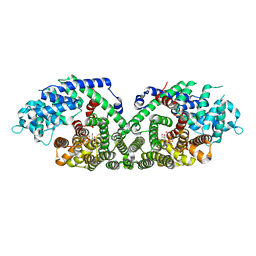 | | (+)-Bornyl Diphosphate Synthase: Complex with Mg and pyrophosphate | | Descriptor: | (+)-bornyl diphosphate synthase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3EKK
 
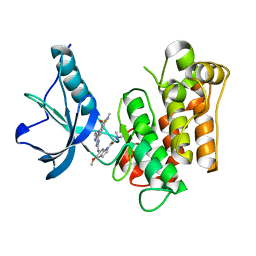 | | Insulin receptor kinase complexed with an inhibitor | | Descriptor: | 2-[(2-{[1-(N,N-dimethylglycyl)-5-methoxy-1H-indol-6-yl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-6-fluoro-N-methylbenzamide, Insulin receptor | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Sabbatini, P, Shewchuk, L. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidines: Potent inhibitors of the IGF-1R receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1MU0
 
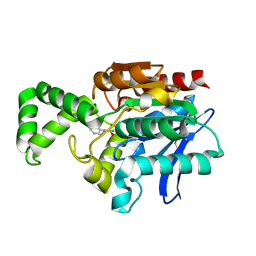 | | Crystal Structure of the Tricorn Interacting Factor F1 Complex with PCK | | Descriptor: | (2R,3S)-3-AMINO-1-CHLORO-4-PHENYL-BUTAN-2-OL, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
8HVC
 
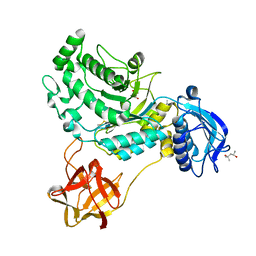 | | Crystal structure of lacto-N-biosidase StrLNBase from Streptomyces sp. strain 142, galacto-N-biose complex 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Fushinobu, S, Yamada, C, Fujio, N. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Glycoside Hydrolase Family 20 Lacto- N -biosidase from Soil Bacterium Streptomyces sp. Strain 142.
J Appl Glycosci (1999), 72, 2025
|
|
5KDU
 
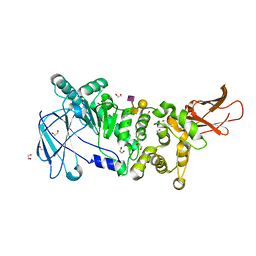 | | ZmpB metallopeptidase in complex with a2,6-Sialyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, SERINE, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3HE8
 
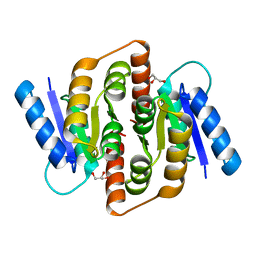 | | Structural study of Clostridium thermocellum Ribose-5-Phosphate Isomerase B | | Descriptor: | GLYCEROL, Ribose-5-phosphate isomerase | | Authors: | Kang, L.W, Kim, J.K, Jung, J.H, Hong, M.K. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
8WDU
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
7FP0
 
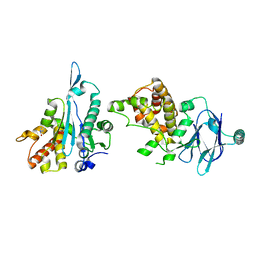 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08F02 from the F2X-Universal Library | | Descriptor: | 1-cyclopropylimidazolidin-2-one, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
6KCD
 
 | |
3HEM
 
 | |
6KCB
 
 | |
5DHJ
 
 | | PIM1 in complex with Cpd4 (3-methyl-5-(pyridin-3-yl)-1H-pyrazolo[3,4-c]pyridine) | | Descriptor: | 3-methyl-5-(pyridin-3-yl)-2H-pyrazolo[3,4-c]pyridine, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H. | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Discovery of 3,5-substituted 6-azaindazoles as potent pan-Pim inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
9LKR
 
 | | Crystal Structure of the bromodomain of human BRD9 in complex with the inhibitor Y22076 | | Descriptor: | 1-[1-[4-(imidazol-1-ylmethyl)-3,5-dimethoxy-phenyl]indolizin-3-yl]ethanone, Bromodomain-containing protein 9, DIMETHYL SULFOXIDE | | Authors: | Chen, Z, Zhang, C, Xu, H, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2025-01-16 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Key imidazolyl groups that induce phenylalanine flipping enhance the efficacy of oral BRD9 inhibitors for AML treatment
Acta Pharm Sin B, 2025
|
|
6Q3Y
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16i | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-(phenylmethyl)-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
5MTK
 
 | | Crystal structure of human Caspase-1 with (3S,6S,10aS)-N-((2S,3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-6-(isoquinoline-1-carboxamido)-5-oxodecahydropyrrolo[1,2-a]azocine-3-carboxamide (PGE-3935199) | | Descriptor: | (3~{S})-3-[[(3~{S},6~{S},10~{a}~{S})-6-(isoquinolin-1-ylcarbonylamino)-5-oxidanylidene-2,3,6,7,8,9,10,10~{a}-octahydro-1~{H}-pyrrolo[1,2-a]azocin-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2017-01-09 | | Release date: | 2018-02-28 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Playing against the odds: scaffold hopping from 3D-fragments
To Be Published
|
|
6EPZ
 
 | |
7BJ6
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-3-(4-chlorophenyl)-4-fluoranyl-3-[[1-(hydroxymethyl)cyclopropyl]methoxy]-6-(2-oxidanylpropan-2-yl)isoindol-1-one, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
8B3L
 
 | | Hen Egg White Lysozyme 2s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
3HIU
 
 | | The crystal structure of protein (XCC3681) from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-20 | | Release date: | 2009-07-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of protein (XCC3681) from Xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
7BIT
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-3-(4-chlorophenyl)-4-fluoranyl-3-[(1-oxidanylcyclopropyl)methoxy]-6-(2-oxidanylpropan-2-yl)isoindol-1-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
