1L3E
 
 | | NMR Structures of the HIF-1alpha CTAD/p300 CH1 Complex | | Descriptor: | ZINC ION, hypoxia inducible factor-1 alpha subunit, p300 protein | | Authors: | Freedman, S.J, Sun, Z.J, Poy, F, Kung, A.L, Livingston, D.M, Wagner, G, Eck, M.J. | | Deposit date: | 2002-02-26 | | Release date: | 2002-04-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
7T03
 
 | | NMR structure of a designed cold unfolding four helix bundle | | Descriptor: | Cold unfolding four helix bundle | | Authors: | Pulavarti, S, Szyperski, T, Yuen, S, Maguire, J, Griffin, J, Kuhlman, B. | | Deposit date: | 2021-11-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
1SF1
 
 | |
2W1O
 
 | | NMR structure of dimerization domain of human ribosomal protein P2 | | Descriptor: | 60S ACIDIC RIBOSOMAL PROTEIN P2 | | Authors: | Lee, K.M, Chan, D.S, Sze, K.H, Zhu, G, Shaw, P.C, Wong, K.B. | | Deposit date: | 2008-10-20 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Dimerization Domain of Ribosomal Protein P2 Provides Insights for the Structural Organization of Eukaryotic Stalk.
Nucleic Acids Res., 38, 2010
|
|
5M9Y
 
 | | NMR solution structure of Harzianin HK-VI in DPC micelles | | Descriptor: | Harzianin HK-VI | | Authors: | Kara, S, Zamora-Carreras, H, Afonin, S, Grage, S.L, Ulrich, A.S, Jimenez, M.A. | | Deposit date: | 2016-11-02 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 11-mer peptaibol Harzianin VI: conformational and biological analysis
To Be Published
|
|
1KKW
 
 | |
1KKV
 
 | |
1RY4
 
 | |
5XEE
 
 | |
1CZ4
 
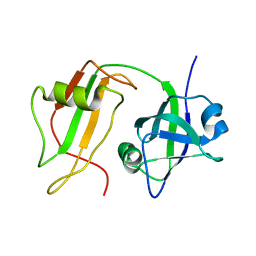 | | NMR STRUCTURE OF VAT-N: THE N-TERMINAL DOMAIN OF VAT (VCP-LIKE ATPASE OF THERMOPLASMA) | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Coles, M, Diercks, T, Liermann, J, Groeger, A, Rockel, B, Baumeister, W, Koretke, K, Lupas, A, Peters, J, Kessler, H. | | Deposit date: | 1999-09-01 | | Release date: | 1999-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of VAT-N reveals a 'missing link' in the evolution of complex enzymes from a simple betaalphabetabeta element.
Curr.Biol., 9, 1999
|
|
2Z4D
 
 | |
1JM4
 
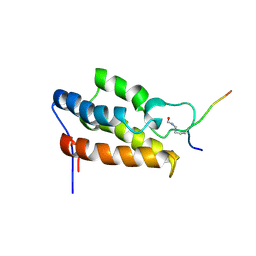 | | NMR Structure of P/CAF Bromodomain in Complex with HIV-1 Tat Peptide | | Descriptor: | HIV-1 Tat Peptide, P300/CBP-associated Factor | | Authors: | Mujtaba, S, He, Y, Zeng, L, Farooq, A, Carlson, J.E, Ott, M, Verdin, E, Zhou, M.-M. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain
Mol.Cell, 9, 2002
|
|
1JJD
 
 | |
3NNH
 
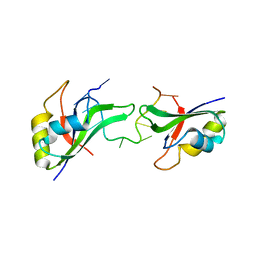 | | Crystal Structure of the CUGBP1 RRM1 with GUUGUUUUGUUU RNA | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7501 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
1P1T
 
 | |
8UWF
 
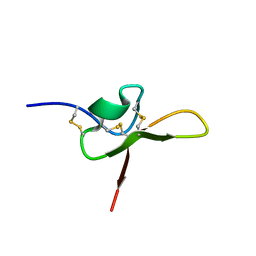 | | NMR structure of the funnel-web spider toxin Hc3a | | Descriptor: | Pi-Hexatoxin-Hc1b_1 | | Authors: | Budusan, E, Payne, C.D, Gonzalez, T.I, Clark, R.J, Rosengren, K.J, Rash, L.D, Cristofori-Armstrong, B. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The funnel-web spider venom derived single knot peptide Hc3a modulates acid-sensing ion channel 1a desensitisation.
Biochem Pharmacol, 2024
|
|
1ZQ3
 
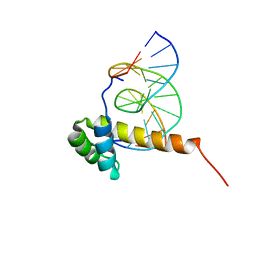 | | NMR Solution Structure of the Bicoid Homeodomain Bound to the Consensus DNA Binding Site TAATCC | | Descriptor: | 5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3', Homeotic bicoid protein | | Authors: | Baird-Titus, J.M, Rance, M, Clark-Baldwin, K, Ma, J, Vrushank, D. | | Deposit date: | 2005-05-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site.
J.Mol.Biol., 356, 2006
|
|
1P9C
 
 | |
6HVC
 
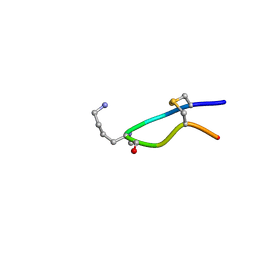 | | NMR structure of Urotensin Peptide Asp-c[Cys-Phe-Trp-(N-Me)Lys-Tyr-Cys]-Val in SDS solution | | Descriptor: | Urotensin-2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Billard, E, Yousif, A.M, Di Maro, S, Abate, L, Bellavita, R, D'Emmanuele di Villa Bianca, R, Santicioli, P, Marinelli, L, Novellino, E, Hebert, T.E, Lubell, W.D, Chatenet, D, Grieco, P. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-16 | | Last modified: | 2019-04-24 | | Method: | SOLUTION NMR | | Cite: | Functional Selectivity Revealed by N-Methylation Scanning of Human Urotensin II and Related Peptides.
J.Med.Chem., 62, 2019
|
|
6HVB
 
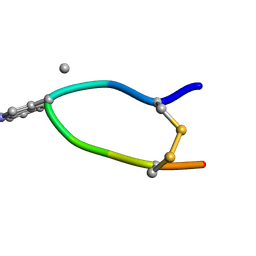 | | NMR structure of Urotensin Peptide Asp-c[Cys-Phe-(N-Me)Trp-Lys-Tyr-Cys]-Val in SDS solution | | Descriptor: | Urotensin-2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Billard, E, Yousif, A.M, Di Maro, S, Abate, L, Bellavita, R, D'Emmanuele di Villa Bianca, R, Santicioli, P, Marinelli, L, Novellino, E, Hebert, T.E, Lubell, W.D, Chatenet, D, Grieco, P. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional Selectivity Revealed by N-Methylation Scanning of Human Urotensin II and Related Peptides.
J.Med.Chem., 62, 2019
|
|
7YF7
 
 | |
6SUU
 
 | |
1L2Y
 
 | |
1IN1
 
 | |
