5XN9
 
 | | Crystal structure of a secretary abundant heat soluble (SAHS) protein from Ramazzottius varieornatus (from monomer sample) | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Fukuda, Y, Miura, Y, Mizohata, E, Inoue, T. | | Deposit date: | 2017-05-19 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural insights into a secretory abundant heat-soluble protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus
FEBS Lett., 591, 2017
|
|
6FZ3
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and inhibitor bound | | Descriptor: | 2,6-bis(chloranyl)-4-[2-(4-methylpiperazin-1-yl)pyridin-4-yl]-~{N}-(1,3,5-trimethylpyrazol-4-yl)benzenesulfonamide, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Kersten, F.C, Brenk, R. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How To Design Selective Ligands for Highly Conserved Binding Sites: A Case Study UsingN-Myristoyltransferases as a Model System.
J.Med.Chem., 63, 2020
|
|
7GRY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-21 | | Descriptor: | 1-(3,5-dichlorophenyl)pyrrolidine-2,5-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
3H3T
 
 | | Crystal structure of the CERT START domain in complex with HPA-16 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]hexadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
7TMU
 
 | | Crystal Structure of the Protein of Unknown Function YPO0625 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function YPO0625 from Yersinia pestis
To Be Published
|
|
7GRX
 
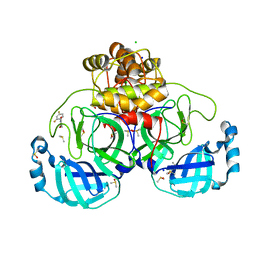 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-20 | | Descriptor: | 1-(2,4-difluorophenyl)pyrrolidine-2,5-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
5I86
 
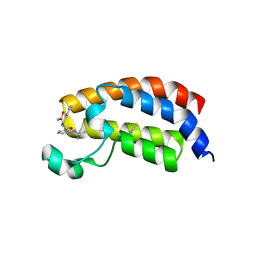 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02778174 | | Descriptor: | (4R)-N-benzyl-4-methyl-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepine-6-carboxamide, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
6P13
 
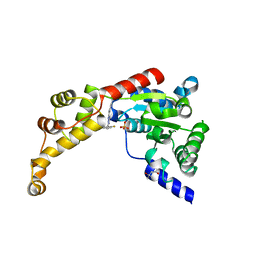 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form A) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, SULFATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
8FD7
 
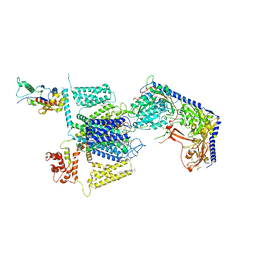 | | Structure of the human L-type voltage-gated calcium channel Cav1.2 complexed with gabapentin | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl heptadecanoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl dodecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Mondal, A, Minor, D.L. | | Deposit date: | 2022-12-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for Ca V alpha 2 delta :gabapentin binding.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5FNG
 
 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 5-[(4-chlorophenyl)methyl]-1,2,3-triaza-4-azanidacyclopenta-2,5-diene, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-11-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
4Q57
 
 | | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Song, J.-G, Kostan, J, Grishkovskaya, I, Djinovic-Carugo, K. | | Deposit date: | 2014-04-16 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex
To be Published
|
|
7H3Y
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1203107138 (A71EV2A-x0517) | | Descriptor: | 1-ethyl-N-[(4-fluorophenyl)methyl]-1H-pyrazole-4-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
9CV4
 
 | |
7JXA
 
 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH INOSITOL 1,4,5-TRIPHOSPHATE | | Descriptor: | 2-ETHOXYETHANOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, S-arrestin, ... | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
6RY9
 
 | |
8RFP
 
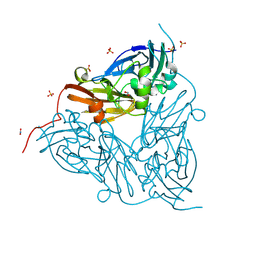 | | Low pH (5.5) nitrite-bound MSOX movie series dataset 3 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [1.83 MGy] - nitrite/NO intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
5X4V
 
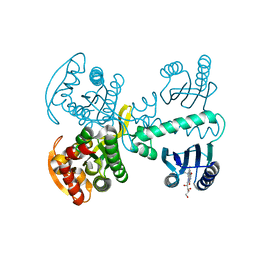 | | Roseoflavin substituted OaPAC | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, Photoactivated adenylyl cyclase | | Authors: | Ohki, M, Park, S.-Y. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of photoactivation of a light-regulated adenylate cyclase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LSQ
 
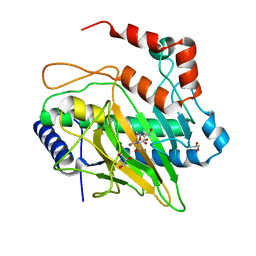 | | Ethylene Forming Enzyme from Pseudomonas syringae pv. phaseolicola - I222 crystal form | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ethylene Forming Enzyme, ... | | Authors: | Zhang, Z, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-09-05 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6DMG
 
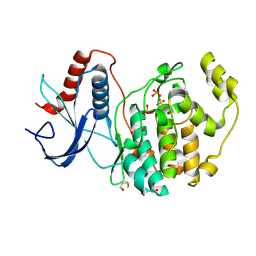 | | A multiconformer ligand model of EK6 bound to ERK2 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Hudson, B.M, van Zundert, G.C.P, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
5I94
 
 | | Crystal structure of human glutaminase C in complex with the inhibitor UPGL-00019 | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}oxy)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R. | | Deposit date: | 2016-02-19 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Design and evaluation of novel glutaminase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
6S03
 
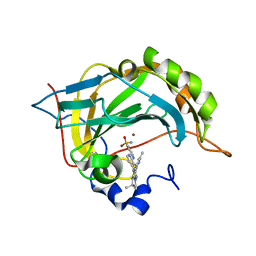 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor I39LT379 | | Descriptor: | 4-[[4-[5,5-dimethyl-2-(6-methylpyridin-2-yl)-4,6-dihydropyrrolo[1,2-b]pyrazol-3-yl]pyridin-2-yl]amino]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
5X7J
 
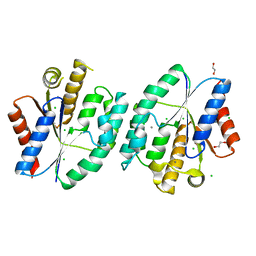 | | Crystal structure of thymidylate kinase from thermus thermophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-02-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insights into product release dynamics through structural analyses of thymidylate kinase.
Int. J. Biol. Macromol., 123, 2019
|
|
7CJ3
 
 | | Crystal structure of the transmembrane domain of Salpingoeca rosetta rhodopsin phosphodiesterase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphodiesterase, RETINAL | | Authors: | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
5BST
 
 | |
5X9A
 
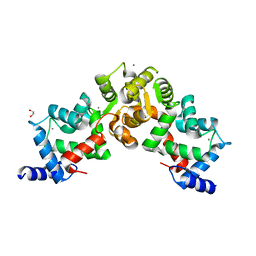 | | Crystal structure of calaxin with calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calaxin | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
