6GOV
 
 | | Structure of THE RNA POLYMERASE LAMBDA-BASED ANTITERMINATION COMPLEX | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, DNA (I), ... | | Authors: | Loll, B, Krupp, F, Said, N, Huang, Y, Buerger, J, Mielke, T, Spahn, C.M.T, Wahl, M.C. | | Deposit date: | 2018-06-04 | | Release date: | 2019-02-13 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for the Action of an All-Purpose Transcription Anti-termination Factor.
Mol.Cell, 74, 2019
|
|
4YT8
 
 | | SeMet-labelled HmdII from Methanocaldococcus jannaschii | | Descriptor: | H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338, TRIETHYLENE GLYCOL | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2015-09-09 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Towards a functional identification of catalytically inactive [Fe]-hydrogenase paralogs.
Febs J., 282, 2015
|
|
4YT5
 
 | | HmdII from Methanocaldococcus jannaschii with bound methylene-tetrahydromethanopterin | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338, TRIETHYLENE GLYCOL | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards a functional identification of catalytically inactive [Fe]-hydrogenase paralogs.
Febs J., 282, 2015
|
|
4REU
 
 | | Revelation of Endogenously bound Fe2+ ions in the Crystal Structure of Ferritin from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FE (III) ION, ... | | Authors: | Thiruselvam, V, Ponnuswamy, M.N, Kumarevel, T.S. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Revelation of endogenously bound Fe(2+) ions in the crystal structure of ferritin from Escherichia coli.
Biochem.Biophys.Res.Commun., 453, 2014
|
|
4EWE
 
 | | Study on structure and function relationships in human Pirin with Manganese ion | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Fu, R, Esaka, S, Chen, L, Serrano, V, Liu, A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1PHZ
 
 | | STRUCTURE OF PHOSPHORYLATED PHENYLALANINE HYDROXYLASE | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
3EYY
 
 | | Structural basis for the specialization of Nur, a nickel-specific Fur homologue, in metal sensing and DNA recognition | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MALONATE ION, ... | | Authors: | Cha, S.-S, An, Y.J. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition
Nucleic Acids Res., 37, 2009
|
|
4TV1
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with propylparaben | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Estrogen receptor, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2014-06-25 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A structural perspective on nuclear receptors as targets of environmental compounds.
Acta Pharmacol.Sin., 36, 2015
|
|
4ERO
 
 | | Study on structure and function relationships in human Pirin with Cobalt ion | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Fu, R, Esaka, S, Chen, L, Serrano, V, Liu, A. | | Deposit date: | 2012-04-20 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6OWN
 
 | |
4EWD
 
 | | Study on structure and function relationships in human Pirin with Mn ion | | Descriptor: | MANGANESE (II) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Chen, L, Fu, R, Serrano, V, Wilson, D.W, Liu, A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8GXF
 
 | | Pseudomonas flexibilis GCN5 family acetyltransferase | | Descriptor: | COENZYME A, GCN5 family acetyltransferase | | Authors: | Song, Y.J, Bao, R. | | Deposit date: | 2022-09-20 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | The novel type II toxin-antitoxin PacTA modulates Pseudomonas aeruginosa iron homeostasis by obstructing the DNA-binding activity of Fur.
Nucleic Acids Res., 50, 2022
|
|
3P63
 
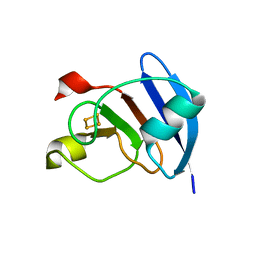 | | Structure of M. laminosus Ferredoxin with a shorter L1,2 loop | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Livnah, O, Nechushtai, R, Eisenberg-Domovich, Y, Michaeli, D. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allostery in the ferredoxin protein motif does not involve a conformational switch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6FCB
 
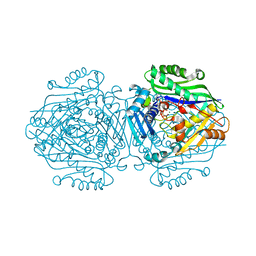 | | Human Methionine Adenosyltransferase II mutant (P115G) | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
6FBP
 
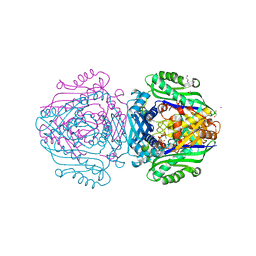 | | Human Methionine Adenosyltransferase II mutant (S114A) in P22121 crystal form | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
3V4Y
 
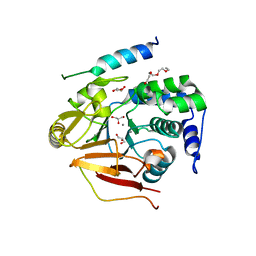 | | Crystal Structure of the first Nuclear PP1 holoenzyme | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Nuclear inhibitor of protein phosphatase 1, ... | | Authors: | Page, R, Peti, W, O'Connell, N.E, Nichols, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The Molecular Basis for Substrate Specificity of the Nuclear NIPP1:PP1 Holoenzyme.
Structure, 20, 2012
|
|
6FBN
 
 | | Human Methionine Adenosyltransferase II mutant (Q113A) | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
8PIL
 
 | | E. coli transcription complex paused at ops site and bound to RfaH and NusA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-22 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
6FBO
 
 | | Human Methionine Adenosyltransferase II mutant (S114A) in I222 crystal form | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, ADENOSINE, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
6FCD
 
 | | Human Methionine Adenosyltransferase II mutant (R264A) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
7VWZ
 
 | | Cryo-EM structure of Rob-dependent transcription activation complex in a unique conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-12 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription activation by Rob, a pleiotropic AraC/XylS family regulator.
Nucleic Acids Res., 50, 2022
|
|
7VWY
 
 | |
5FMF
 
 | | the P-lobe of RNA polymerase II pre-initiation complex | | Descriptor: | DNA REPAIR HELICASE RAD25, SSL2, DNA REPAIR HELICASE RAD3, ... | | Authors: | Murakami, K, Tsai, K, Kalisman, N, Bushnell, D.A, Asturias, F.J, Kornberg, R.D. | | Deposit date: | 2015-11-03 | | Release date: | 2015-11-25 | | Last modified: | 2017-07-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of an RNA Polymerase II Pre-Initiation Complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8PID
 
 | | backtracked E. coli transcription complex paused at ops site and bound to RfaH | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-21 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
8PFJ
 
 | | fully recruited RfaH bound to E. coli transcription complex paused at ops site (not fully complementary scaffold; alternative state of RfaH) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
