5T9P
 
 | |
5EN1
 
 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*GP*GP*AP*CP*UP*G)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2015-11-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1
Nat Commun, 2018
|
|
5IFM
 
 | | Human NONO (p54nrb) Homodimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-POU domain-containing octamer-binding protein, ... | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of human NONO (p54(nrb)): overcoming pathological problems with purification, data collection and noncrystallographic symmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5LSO
 
 | |
2N7C
 
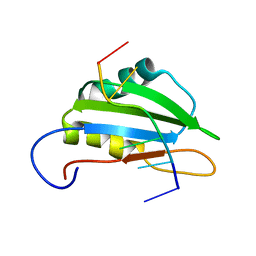 | |
5LQW
 
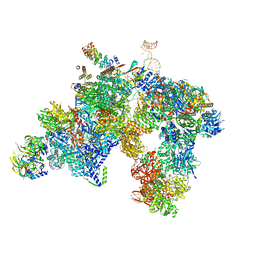 | | yeast activated spliceosome | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-processing protein 45, Pre-mRNA-splicing factor 8, ... | | Authors: | Rauhut, R, Luehrmann, R. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Molecular architecture of the Saccharomyces cerevisiae activated spliceosome
Science, 6306, 2016
|
|
5GM6
 
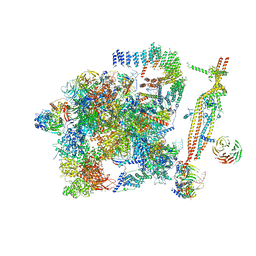 | | Cryo-EM structure of the activated spliceosome (Bact complex) at 3.5 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cold sensitive U2 snRNA suppressor 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a yeast activated spliceosome at 3.5 angstrom resolution
Science, 353, 2016
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | Descriptor: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
5GMK
 
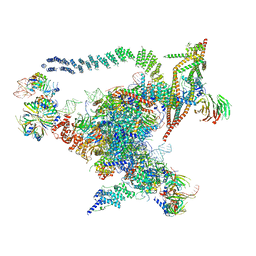 | | Cryo-EM structure of the Catalytic Step I spliceosome (C complex) at 3.4 angstrom resolution | | Descriptor: | 5'-Exon, 5'-Splicing Site, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wan, R, Yan, C, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a yeast catalytic step I spliceosome at 3.4 angstrom resolution
Science, 353, 2016
|
|
5D77
 
 | | Structure of Mip6 RRM3 Domain | | Descriptor: | CITRIC ACID, NITRATE ION, RNA-binding protein MIP6, ... | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2015-08-13 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Mip6 RRM3 domain at 1.3
To Be Published
|
|
5D78
 
 | |
5LJ3
 
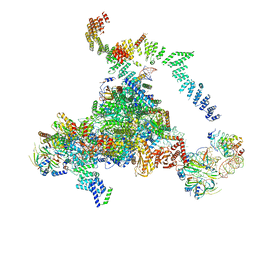 | | Structure of the core of the yeast spliceosome immediately after branching | | Descriptor: | CEF1, CLF1, CWC15, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-03 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
5IM0
 
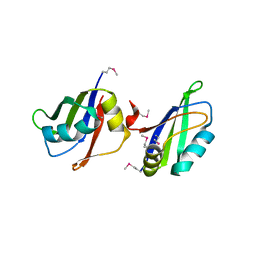 | |
2N3L
 
 | |
5K1H
 
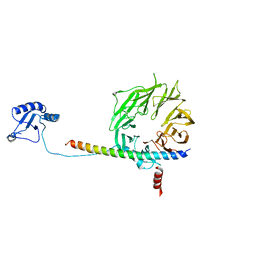 | | eIF3b relocated to the intersubunit face to interact with eIF1 and below the eIF2 ternary-complex. from the structure of a partial yeast 48S preinitiation complex in closed conformation. | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, eIF3a C-terminal tail | | Authors: | Simonetti, A, Brito Querido, J, Myasnikov, A.G, Mancera-Martinez, E, Renaud, A, Kuhn, L, Hashem, Y. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition.
Mol.Cell, 63, 2016
|
|
5K0Y
 
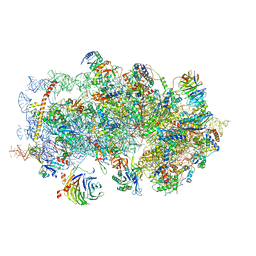 | | m48S late-stage initiation complex, purified from rabbit reticulocytes lysates, displaying eIF2 ternary complex and eIF3 i and g subunits relocated to the intersubunit face | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S12, 40S ribosomal protein S21, ... | | Authors: | Simonetti, A, Brito Querido, J, Myasnikov, A.G, Mancera-Martinez, E, Renaud, A, Kuhn, L, Hashem, Y. | | Deposit date: | 2016-05-17 | | Release date: | 2016-07-13 | | Last modified: | 2018-04-18 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition.
Mol.Cell, 63, 2016
|
|
4ZKA
 
 | | High Resolution Crystal Structure of Fox1 RRM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, RNA binding protein fox-1 homolog 1, SULFATE ION, ... | | Authors: | Blatter, M, Allain, F.H.-T. | | Deposit date: | 2015-04-30 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Resolution Crystal Structure of Fox1 RRM
To Be Published
|
|
3JCT
 
 | | Cryo-em structure of eukaryotic pre-60S ribosomal subunits | | Descriptor: | 60S ribosomal protein L11-A, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Wu, S, Kumcuoglu, B, Yan, K.G, Brown, H, Zhang, Y.X, Tan, D, Gamalinda, M, Yuan, Y, Li, Z.F, Jakovljevic, J, Ma, C.Y, Lei, J.L, Dong, M.Q, Woolford Jr, J.L, Gao, N. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes
Nature, 534, 2016
|
|
5FJ4
 
 | |
5IQQ
 
 | |
2N82
 
 | |
5EV1
 
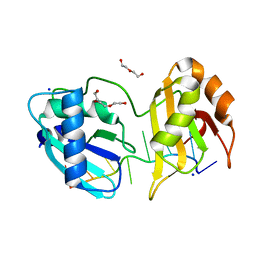 | | Structure I of Intact U2AF65 Recognizing a 3' Splice Site Signal | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(*UP*UP*U)-D(P*UP*UP*(BRU)P*U)-R(P*UP*U)-3'), SODIUM ION, ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
5EV2
 
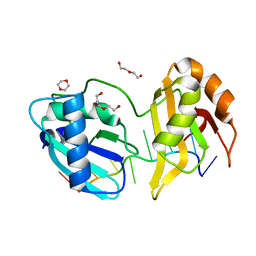 | | Structure II of Intact U2AF65 Recognizing the 3' Splice Site Signal | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DI(HYDROXYETHYL)ETHER, DNA (5'-R(P*UP*U)-D(P*UP*U)-R(P*U)-D(P*UP*(BRU)P*U)-3'), ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
5EV3
 
 | |
5EV4
 
 | |
