8DID
 
 | |
8DIC
 
 | |
8DIB
 
 | |
8CA9
 
 | |
8BF9
 
 | | Molecular view of ER membrane remodeling by the Sec61/TRAP translocon. | | Descriptor: | 60S ribosomal protein L39, Large ribosomal subunit protein eL31, Large ribosomal subunit protein eL38, ... | | Authors: | Karki, S, Javanainen, M, Tranter, D, Rehan, S, Huiskonen, J, Happonen, L, Paavilainen, V. | | Deposit date: | 2022-10-24 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Molecular view of ER membrane remodeling by the Sec61/TRAP translocon.
Embo Rep., 24, 2023
|
|
8AZK
 
 | | Bovine 20S proteasome, untreated | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Szenkier, N, Arie, M, Matzov, D, Sertchook, R, Carmeli, R, Cascio, P, Stanhill, A, Shalev Benami, M, Navon, A. | | Deposit date: | 2022-09-06 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bovine 20S proteasome, untreated
To be published
|
|
7ZL3
 
 | |
7ZEB
 
 | | Complex I from Ovis aries at pH9, Closed state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-30 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZDP
 
 | | Complex I from Ovis aries at pH9, Open state | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZDM
 
 | | Complex I from Ovis aries at pH5.5, Closed state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZDJ
 
 | | Complex I from Ovis aries at pH5.5, Open state | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Acyl carrier protein, Complex I subunit B13, ... | | Authors: | Petrova, O, Sazanov, L. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZDH
 
 | | Complex I from Ovis aries at pH7.4, Closed state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZD6
 
 | | Complex I from Ovis aries, at pH7.4, Open state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z0G
 
 | | CPAP:TUBULIN:IE5 ALPHAREP COMPLEX P1 SPACE GROUP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Centromere protein J, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z0F
 
 | | CPAP:S-TUBULIN:IIH5 ALPHAREP COMPLEX | | Descriptor: | Centromere protein J, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YOB
 
 | |
7WUO
 
 | |
7VE3
 
 | | Structure of the complex of sheep lactoperoxidase with hypoiodite at 2.70 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, P.K, Yamini, S, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence of the oxidation of iodide ion into hyper-reactive hypoiodite ion by mammalian heme lactoperoxidase.
Protein Sci., 31, 2022
|
|
7Q1F
 
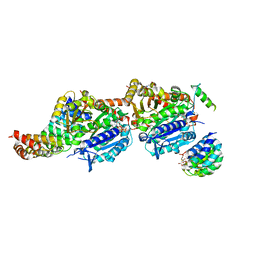 | | CPAP:TUBULIN:IE5 ALPHAREP COMPLEX | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CITRATE ANION, Centromere protein J, ... | | Authors: | Campanacci, V, Gigant, b. | | Deposit date: | 2021-10-19 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q1E
 
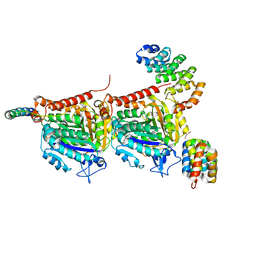 | | CPAP:TUBULIN:IIH5 ALPHAREP COMPLEX | | Descriptor: | Centromere protein J, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Campanacci, V, Gigant, b. | | Deposit date: | 2021-10-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ONI
 
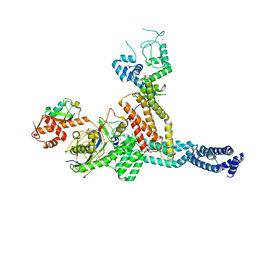 | | Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* | | Descriptor: | Cullin-5, E3 ubiquitin-protein ligase ARIH2, NEDD8, ... | | Authors: | Kostrhon, S.P, prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation.
Nat.Chem.Biol., 17, 2021
|
|
7OD1
 
 | |
7NF8
 
 | | Ovine (b0,+AT-rBAT)2 hetero-tetramer, asymmetric unit, rigid-body fitted | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7NF7
 
 | | Ovine rBAT ectodomain homodimer, asymmetric unit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7NF6
 
 | | Ovine b0,+AT-rBAT heterodimer | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
