2LAS
 
 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | Descriptor: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | Authors: | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | Deposit date: | 2011-03-20 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
2IYD
 
 | | SENP1 covalent complex with SUMO-2 | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 1, SMALL UBIQUITIN-RELATED MODIFIER 2 | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-14 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Senp1 Native Structure
To be Published
|
|
2IO3
 
 | | Crystal structure of human Senp2 in complex with RanGAP1-SUMO-2 | | Descriptor: | Ran GTPase-activating protein 1, Sentrin-specific protease 2, Small ubiquitin-related modifier 2 | | Authors: | Reverter, D, Lima, C.D. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4BKG
 
 | | crystal structure of human diSUMO-2 | | Descriptor: | SMALL UBIQUITIN-RELATED MODIFIER 2 | | Authors: | Keusekotten, K, Bade, V.N, Meyer-Teschendorf, K, Sriramachandran, A, Fischer-Schrader, K, Krause, A, Horst, C, Hofmann, K, Dohmen, R.J, Praefcke, G.J.K. | | Deposit date: | 2013-04-25 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Multivalent Interactions of the Sumo-Interaction Motifs in the Ring-Finger Protein 4 (Rnf4) Determine the Specificity for Chains of the Small Ubiquitin-Related Modifier (Sumo).
Biochem.J., 457, 2014
|
|
3UIP
 
 | | Complex between human RanGAP1-SUMO1, UBC9 and the IR1 domain from RanBP2 containing IR2 Motif II | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
2IO0
 
 | | Crystal structure of human Senp2 in complex with preSUMO-2 | | Descriptor: | SULFATE ION, Sentrin-specific protease 2, Small ubiquitin-related modifier 2 precursor | | Authors: | Reverter, D, Lima, C.D. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates.
Nat.Struct.Mol.Biol., 13, 2006
|
|
6K5R
 
 | | Complex of SUMO2 with Phosphorylated viral SIM IE2 | | Descriptor: | ASP-THR-ALA-GLY-CYS-ILE-VAL-ILE-SEP-ASP-SEP-GLU, Small ubiquitin-related modifier 3 | | Authors: | Chatterjee, K.S, Das, R. | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Casein kinase-2-mediated phosphorylation increases the SUMO-dependent activity of the cytomegalovirus transactivator IE2.
J.Biol.Chem., 294, 2019
|
|
5EQL
 
 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SUMO-Affirmer-S2D5, Small ubiquitin-related modifier 2 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
5ELU
 
 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SULFATE ION, SUMO-Affirmer-S2B3, Small ubiquitin-related modifier 2 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-05 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
8ITY
 
 | | human RNA polymerase III pre-initiation complex closed DNA 1 | | Descriptor: | DNA (82-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
6RYU
 
 | | Nucleosome-CHD4 complex structure (two CHD4 copies) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4,CHD4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | Authors: | Farnung, L, Ochmann, M, Cramer, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
6RYR
 
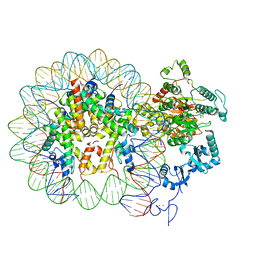 | | Nucleosome-CHD4 complex structure (single CHD4 copy) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | Authors: | Farnung, L, Ochmann, M, Cramer, P. | | Deposit date: | 2019-06-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
7NFX
 
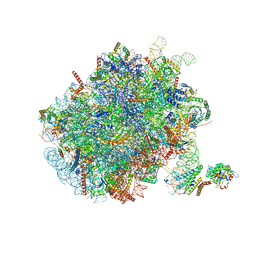 | | Mammalian ribosome nascent chain complex with SRP and SRP receptor in early state A | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Lee, J.H, Shan, S, Ban, N. | | Deposit date: | 2021-02-08 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER.
Sci Adv, 7, 2021
|
|
4V6X
 
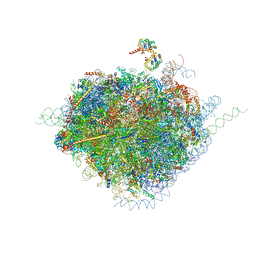 | | Structure of the human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
7DN3
 
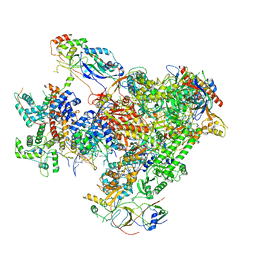 | | Structure of Human RNA Polymerase III elongation complex | | Descriptor: | DNA (5'-D(P*TP*CP*GP*TP*CP*TP*GP*AP*TP*CP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*CP*GP*AP*GP*AP*TP*CP*AP*GP*AP*CP*GP*AP*GP*AP*TP*CP*GP*GP*G)-3'), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Li, L, Yu, Z, Zhao, D, Ren, Y, Hou, H, Xu, Y. | | Deposit date: | 2020-12-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of human RNA polymerase III elongation complex.
Cell Res., 31, 2021
|
|
7D58
 
 | | cryo-EM structure of human RNA polymerase III in elongating state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D59
 
 | | cryo-EM structure of human RNA polymerase III in apo state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5WWF
 
 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
8C60
 
 | | Cryo-EM structure of the human SIN3B full-length complex at 3.4 Angstrom resolution | | Descriptor: | CALCIUM ION, Histone deacetylase 2, Isoform 2 of Paired amphipathic helix protein Sin3b, ... | | Authors: | Alfieri, C, Wan, S.M, Muhammad, R. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of assembly, activation and lysine selection by the SIN3B histone deacetylase complex.
Nat Commun, 14, 2023
|
|
1EJ9
 
 | | CRYSTAL STRUCTURE OF HUMAN TOPOISOMERASE I DNA COMPLEX | | Descriptor: | DNA (5'-D(*C*AP*AP*AP*AP*AP*GP*AP*CP*TP*CP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*C*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*GP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), DNA TOPOISOMERASE I | | Authors: | Redinbo, M.R, Champoux, J.J, Hol, W.G. | | Deposit date: | 2000-03-01 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA.
Biochemistry, 39, 2000
|
|
6R7Q
 
 | | Structure of XBP1u-paused ribosome nascent chain complex with Sec61. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shanmuganathan, V, Cheng, J, Braunger, K, Berninghausen, O, Beatrix, B, Beckmann, R. | | Deposit date: | 2019-03-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
6R8Y
 
 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
7YQ8
 
 | | Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, 50-mer DNA, DNA topoisomerase 2-beta, ... | | Authors: | Naganuma, M, Ehara, H, Kim, D, Nakagawa, R, Cong, A, Bu, H, Jeong, J, Jang, J, Schellenberg, M.J, Bunch, H, Sekine, S. | | Deposit date: | 2022-08-05 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ERK2-topoisomerase II regulatory axis is important for gene activation in immediate early genes.
Nat Commun, 14, 2023
|
|
2GRO
 
 | | Crystal Structure of human RanGAP1-Ubc9-N85Q | | Descriptor: | Ran GTPase-activating protein 1, Ubiquitin-conjugating enzyme E2 I | | Authors: | Yunus, A.A, Lima, C.D. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2GRN
 
 | | Crystal Structure of human RanGAP1-Ubc9 | | Descriptor: | Ran GTPase-activating protein 1, Ubiquitin-conjugating enzyme E2 I | | Authors: | Yunus, A.A, Lima, C.D. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway.
Nat.Struct.Mol.Biol., 13, 2006
|
|
