4TU0
 
 | | CRYSTAL STRUCTURE OF CHIKUNGUNYA VIRUS NSP3 MACRO DOMAIN IN COMPLEX WITH A 2'-5' OLIGOADENYLATE TRIMER | | Descriptor: | 2'-5' OLIGOADENYLATE TRIMER, DI(HYDROXYETHYL)ETHER, Non-structural polyprotein 3 | | Authors: | Morin, B, Ferron, f.p, Malet, h, Coutard, b, Canard, b. | | Deposit date: | 2014-06-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF CHIKUNGUNYA VIRUS NSP3 MACRO DOMAIN IN COMPLEX WITH A 2'-5' OLIGOADENYLATE TRIMER
TO BE PUBLISHED
|
|
3Q6Z
 
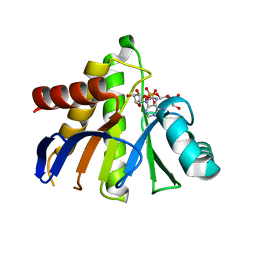 | | HUman PARP14 (ARTD8)-Macro domain 1 in complex with adenosine-5-diphosphoribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Thorsell, A.G, Tresaugues, L, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
3Q71
 
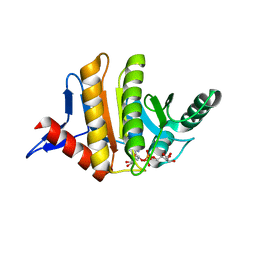 | | Human parp14 (artd8) - macro domain 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Thorsell, A.G, Tresaugues, L, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
4UML
 
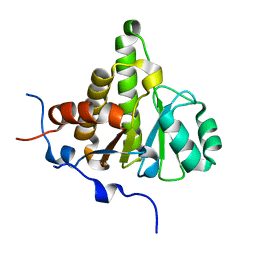 | | Crystal structure of ganglioside induced differentiation associated protein 2 (GDAP2) macro domain | | Descriptor: | GANGLIOSIDE-INDUCED DIFFERENTIATION-ASSOCIATED PROTEIN 2 | | Authors: | Elkins, J.M, Wang, J, Kopec, J, Wang, D, Strain-Damerell, C, Shrestha, L, Sieg, C, Tallant, C, Newman, J.A, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2014-05-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Gdap2
To be Published
|
|
3VFQ
 
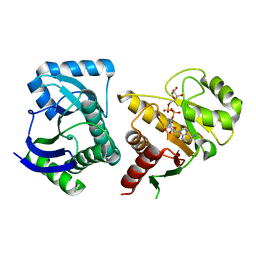 | | Human PARP14 (ARTD8, BAL2) - macro domains 1 and 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
6W7H
 
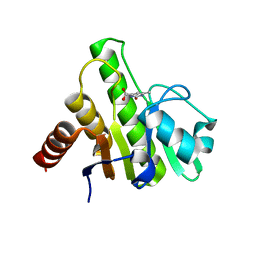 | | Co-crystal structures of CHIKV nsP3 macrodomain with pyrimidone fragments | | Descriptor: | 6-methyl-2-oxo-2,5-dihydropyrimidine-4-carboxylic acid, Nonstructural polyprotein | | Authors: | Zhang, S, Garzan, A, Augelli-Szafran, C.E, Pathak, A.K, Wu, M. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6W0T
 
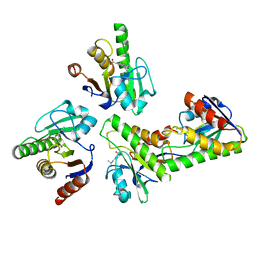 | |
7NY7
 
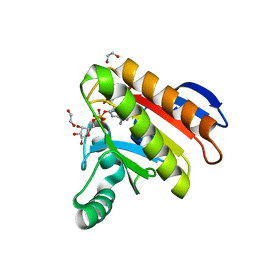 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Histone macroH2A1.1 | | Authors: | Guberovic, I, Knobloch, G, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NY6
 
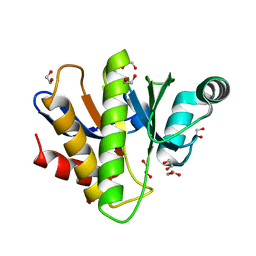 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Histone macroH2A1.1 | | Authors: | Knobloch, G, Guberovic, I, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4IQY
 
 | | Crystal structure of the human protein-proximal ADP-ribosyl-hydrolase MacroD2 | | Descriptor: | MAGNESIUM ION, O-acetyl-ADP-ribose deacetylase MACROD2, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Jankevicius, G, Hassler, M, Golia, B, Rybin, V, Zacharias, M, Timinszky, G, Ladurner, A.G. | | Deposit date: | 2013-01-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A family of macrodomain proteins reverses cellular mono-ADP-ribosylation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4J5S
 
 | | TARG1 (C6orf130), Terminal ADP-ribose Glycohydrolase 1 ADP-ribose complex | | Descriptor: | 1,2-ETHANEDIOL, 1,3,2-DIOXABOROLAN-2-OL, BORATE ION, ... | | Authors: | Schellenberg, M.J, Appel, C.D, Krahn, J, Williams, R.S. | | Deposit date: | 2013-02-09 | | Release date: | 2013-03-27 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Embo J., 32, 2013
|
|
9AZX
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with NDPr | | Descriptor: | Non-structural protein 3, {(2R,3S,4R,5R)-5-[(8S)-4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl]-5-cyano-3,4-dihydroxyoxolan-2-yl}methyl [(2R,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | GS-441524-Diphosphate-Ribose Derivatives as Nanomolar Binders and Fluorescence Polarization Tracers for SARS-CoV-2 and Other Viral Macrodomains.
Acs Chem.Biol., 19, 2024
|
|
5ZUB
 
 | |
7P27
 
 | | NMR solution structure of Chikungunya virus macro domain | | Descriptor: | Polyprotein P1234 | | Authors: | Lykouras, M.V, Tsika, A.C, Papageorgiou, N, Canard, B, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2021-07-04 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
1VHU
 
 | | Crystal structure of a putative phosphoesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypothetical protein AF1521 | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
5ZUA
 
 | |
8TV6
 
 | | SARS-CoV-2 Mac1 in complex with MDOLL-0169 | | Descriptor: | (1R,6R)-6-{[3-(methoxycarbonyl)-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophen-2-yl]carbamoyl}cyclohex-3-ene-1-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wazir, S, Maksimainen, M, Lehtio, L. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of 2-Amide-3-methylester Thiophenes that Target SARS-CoV-2 Mac1 and Repress Coronavirus Replication, Validating Mac1 as an Antiviral Target.
J.Med.Chem., 67, 2024
|
|
8TV7
 
 | | SARS-CoV-2 Mac1 in complex with MDOLL-0229 | | Descriptor: | (1R,2R)-2-{[3-(methoxycarbonyl)-4,5,6,7,8,9-hexahydrocycloocta[b]thiophen-2-yl]carbamoyl}cyclohexane-1-carboxylic acid, GLYCEROL, Papain-like protease nsp3 | | Authors: | Wazir, S, Maksimainen, M, Lehtio, L. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 2-Amide-3-methylester Thiophenes that Target SARS-CoV-2 Mac1 and Repress Coronavirus Replication, Validating Mac1 as an Antiviral Target.
J.Med.Chem., 67, 2024
|
|
4J5R
 
 | | TARG1 (C6orf130), Terminal ADP-ribose Glycohydrolase 1 bound to ADP-HPD | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, CHLORIDE ION, ... | | Authors: | Schellenberg, M.J, Appel, C.D, Krahn, J, Williams, R.S. | | Deposit date: | 2013-02-09 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Embo J., 32, 2013
|
|
8SH8
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain Asn40Asp mutant in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Papain-like protease nsp3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2023-04-13 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal structure of SARS-CoV-2 NSP3 macrodomain Asn40Asp mutant in complex with ADP-ribose (P43 crystal form)
To Be Published
|
|
8SH6
 
 | |
8P0C
 
 | | Rubella virus p150 macro domain (apo) | | Descriptor: | Non-structural polyprotein p200 | | Authors: | Stoll, G.A, Modis, Y. | | Deposit date: | 2023-05-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure and biochemical activity of the macrodomain from rubella virus p150.
J.Virol., 98, 2024
|
|
8P0E
 
 | |
5ZU7
 
 | |
