9J4P
 
 | | Regulatory domain and kinase domain of ALPK1 protein | | Descriptor: | Alpha-protein kinase 1, ZINC ION, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5S,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Xu, C, Xu, T. | | Deposit date: | 2024-08-09 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Regulatory domain and kinase domain of ALPK1 protein
To Be Published
|
|
4QGX
 
 | | Crystal structure of the R132K:R111L:L121E mutant of Cellular Retinoic Acid Binding ProteinII complexed with a synthetic ligand (Merocyanine) at 1.47 angstrom resolution | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, Cellular retinoic acid-binding protein 2 | | Authors: | Nosrati, M, Yapici, I, Geiger, J.H. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
7TXT
 
 | | Structure of human serotonin transporter bound to small molecule '8090 in lipid nanodisc and NaCl | | Descriptor: | 1-[4-(4-fluorophenyl)-1,3-thiazol-2-yl]piperazine, 15B8 Fab heavy chain, 15B8 Fab light chain, ... | | Authors: | Singh, I, Seth, A, Billesboelle, C.B, Braz, J, Rodriguiz, R.M, Roy, K, Bekele, B, Craik, V, Huang, X.P, Boytsov, D, Lak, P, O'Donnell, H, Sandtner, W, Roth, B.L, Basbaum, A.I, Wetsel, W.C, Manglik, A, Shoichet, B.K, Rudnick, G. | | Deposit date: | 2022-02-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based discovery of conformationally selective inhibitors of the serotonin transporter.
Cell, 186, 2023
|
|
7L6L
 
 | | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Deoxyribose operon repressor, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12.
To Be Published
|
|
7L75
 
 | | Crystal Structure of Peptidylprolyl Isomerase PrsA from Streptococcus mutans. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Foldase protein PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure of Peptidylprolyl Isomerase PrsA from Streptococcus mutans.
To Be Published
|
|
4XZB
 
 | |
4QEF
 
 | | Human Carbonic Anhydrase II V207I - cyanate inhibitor complex | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION, ... | | Authors: | West, D.M. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Human carbonic anhydrase II-cyanate inhibitor complex: putting the debate to rest.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
9DNM
 
 | | Structure of rat beta-arrestin 1 bound to allosteric inhibitor | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, Beta-arrestin-1,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | Authors: | Pakharukova, N, Kahsai, A.W, Masoudi, A, Lefkowitz, R.J. | | Deposit date: | 2024-09-17 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Small-molecule modulation of beta-arrestins.
Biorxiv, 2025
|
|
8OXA
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2-Pi conformation with occluded PS | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
6SQY
 
 | | Mouse dCTPase in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
4QAH
 
 | | The second sphere residue T263 is important for function and activity of PTP1B through modulating WPD loop | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xiao, P, Wang, X, Wang, H.M, Fu, X.L, Cui, F.A, Bi, W.X. | | Deposit date: | 2014-05-05 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | The second-sphere residue T263 is important for the function and catalytic activity of PTP1B via interaction with the WPD-loop
Int.J.Biochem.Cell Biol., 57C, 2014
|
|
1MDX
 
 | | Crystal structure of ArnB transferase with pyridoxal 5' phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T.A, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
6E4F
 
 | | Crystal structure of ARQ 531 in complex with the kinase domain of BTK | | Descriptor: | 1,2-ETHANEDIOL, 1,5-anhydro-2-{[5-(2-chloro-4-phenoxybenzene-1-carbonyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}-2,3,4-trideoxy-D-erythro-hexitol, IMIDAZOLE, ... | | Authors: | Eathiraj, S. | | Deposit date: | 2018-07-17 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The BTK Inhibitor ARQ 531 Targets Ibrutinib-Resistant CLL and Richter Transformation.
Cancer Discov, 8, 2018
|
|
6PEG
 
 | | MIF with a allosteric inhibitor | | Descriptor: | 4-amino-5-hydroxy-6-[(E)-(3-{[3-(2-methylpropanoyl)pyrazolo[1,5-a]pyridin-2-yl]methyl}phenyl)diazenyl]naphthalene-1,3-disulfonic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of Macrophage Migration Inhibitory Factor by a Chimera of Two Allosteric Binders.
Acs Med.Chem.Lett., 11, 2020
|
|
5JDK
 
 | |
5ZXM
 
 | | Crystal Structure of GyraseB N-terminal at 1.93A Resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA gyrase subunit B, ... | | Authors: | Tiwari, P, Gupta, D, Sachdeva, E, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Structural insights into the transient closed conformation and pH dependent ATPase activity of S.Typhi GyraseB N- terminal domain.
Arch.Biochem.Biophys., 701, 2021
|
|
7YC3
 
 | | Crystal structure of FGFR4 kinase domain with 10t | | Descriptor: | 6-bromanyl-~{N}-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-5-methanoyl-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide, Fibroblast growth factor receptor 4, GLYCEROL, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC1
 
 | | Crystal structure of FGFR4 kinase domain with 10d | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.535 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
5C6C
 
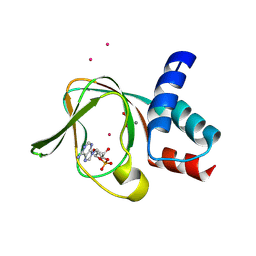 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cAMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CADMIUM ION, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-22 | | Release date: | 2016-01-20 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
7A0S
 
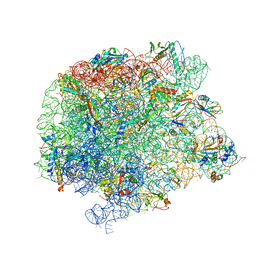 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
7KTF
 
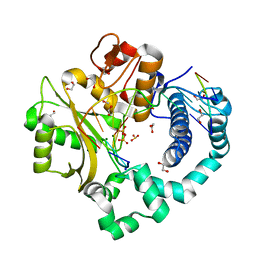 | | DNA Polymerase Mu, 8-oxodGTP:Ct Product State Ternary Complex, 50 mM Mg2+ (180min) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*CP*GP*GP*CP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
9GCJ
 
 | |
5J59
 
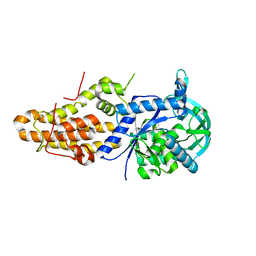 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor (Chem 1893) | | Descriptor: | 5-chloro-N-(2-{[(3,5-dichlorophenyl)methyl]amino}ethyl)-N-methyl-3H-imidazo[4,5-b]pyridin-2-amine, GLYCEROL, METHIONINE, ... | | Authors: | Barros-Alvarez, X, Hol, W.G.J. | | Deposit date: | 2016-04-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided design of novel Trypanosoma brucei Methionyl-tRNA synthetase inhibitors.
Eur J Med Chem, 124, 2016
|
|
9JSE
 
 | | Crystal Structure of PenI beta-Lactamase from Burkholderia pseudomallei Complex with Taniborbactam | | Descriptor: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Beta-lactamase, GLYCEROL | | Authors: | Nukaga, M, Hoshino, T, Mojica, M.F, Papp-Wallace, K.M. | | Deposit date: | 2024-09-30 | | Release date: | 2025-08-13 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Burkholderia pseudomallei PenI beta-lactamase and variants are potently inhibited by taniborbactam.
Antimicrob.Agents Chemother., 69, 2025
|
|
8OXB
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2-Pi conformation with occluded PC | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
