9JBH
 
 | | Cryo-EM structure of the human LYCHOS PLD homodimer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, Lysosomal cholesterol signaling protein | | Authors: | Yu, S, Liang, L. | | Deposit date: | 2024-08-27 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural insights into cholesterol sensing by the LYCHOS-mTORC1 pathway.
Nat Commun, 16, 2025
|
|
4TYI
 
 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, Fibroblast growth factor receptor 4 | | Authors: | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
9FF7
 
 | | Structure of the BMOE-crosslinked transcription termination factor Rho in the presence of ppGpp; S84C/M405C double mutant | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), MAGNESIUM ION, Transcription termination factor Rho | | Authors: | Said, N, Hilal, T, Wahl, M.C. | | Deposit date: | 2024-05-22 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nucleotide-induced hyper-oligomerization inactivates transcription termination factor rho.
Nat Commun, 16, 2025
|
|
4TYL
 
 | | Fragment-Based Screening of the Bromodomain of ATAD2 | | Descriptor: | 5-amino-1,3,6-trimethyl-1,3-dihydro-2H-benzimidazol-2-one, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-08 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
7ZWS
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 13 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-cyano-6-thiophen-2-yl-4-(trifluoromethyl)pyridin-2-yl]sulfanyl-2-phenyl-ethanoic acid, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
4TUT
 
 | | Structure of a Prion peptide | | Descriptor: | Prion peptide: GLY-GLY-TYR-MET-LEU-GLY | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-06-24 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
9MKC
 
 | | Crystal structure of MALT1 in complex with an allosteric inhibitor | | Descriptor: | CALCIUM ION, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, N-[(8R)-2-chloro-7-(propan-2-yl)pyrazolo[1,5-a]pyrimidin-6-yl]-N'-[5-chloro-6-(2H-1,2,3-triazol-2-yl)pyridin-3-yl]urea | | Authors: | Bell, J.A. | | Deposit date: | 2024-12-17 | | Release date: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Accelerated In Silico Discovery of SGR-1505 : A Potent MALT1 Allosteric Inhibitor for the Treatment of Mature B-Cell Malignancies.
J.Med.Chem., 2025
|
|
7ZWR
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-[[(6~{S})-3-cyano-6-methyl-4-(trifluoromethyl)-5,6,7,8-tetrahydroquinolin-2-yl]sulfanyl]ethanoylamino]ethanoic acid, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
6I5L
 
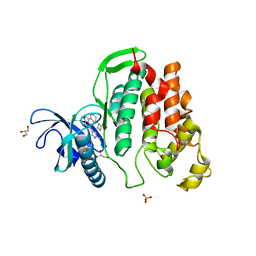 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound VN316 (derivative of compound 12h) | | Descriptor: | 3-(3-cyclobutylphenyl)-5-(1-methylpyrazol-4-yl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
7ZWZ
 
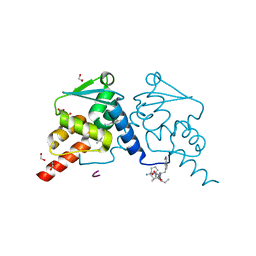 | | Crystal structure of human BCL6 BTB domain in complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, B-cell lymphoma 6 protein, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
6LLA
 
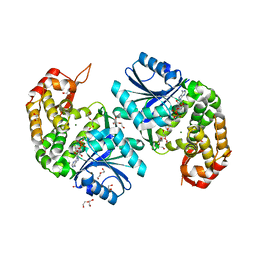 | | Crystal structure of Providencia alcalifaciens 3-dehydroquinate synthase (DHQS) in complex with Mg2+ and NAD | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Neetu, N, Katiki, M, Kumar, P. | | Deposit date: | 2019-12-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and Biochemical Analyses Reveal that Chlorogenic Acid Inhibits the Shikimate Pathway.
J.Bacteriol., 202, 2020
|
|
5NRI
 
 | |
9F8M
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with [6-Hydroxy-2-(4-hydroxyphenyl)benzo[b]thiophen-3-yl][4-(propylamino)phenyl]methanone | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Phenazine biosynthesis protein A/B, ... | | Authors: | Zimmermann, M, Thiemann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
4UD9
 
 | | Thrombin in complex with 5-chlorothiophene-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-CHLORO-2-THIOPHENECARBOXAMIDE, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2014-12-09 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
5BOE
 
 | | Crystal structure of Staphylococcus aureus enolase in complex with PEP | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wang, C.L, Wu, Y.F, Han, L, Wu, M.H, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6G1D
 
 | | Corynebacterium glutamicum OxyR C206 mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Young, D.R, Pedre, B.P, Messens, J.M. | | Deposit date: | 2018-03-21 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7XO9
 
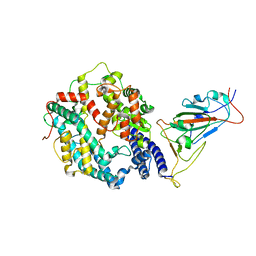 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
5Z1M
 
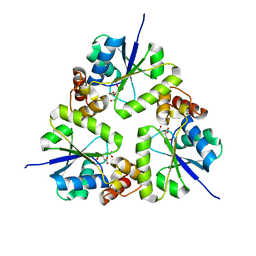 | | Crystal structure of the complex of trimeric Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with citrate ion at 1.87 A resolution | | Descriptor: | CITRIC ACID, Phosphopantetheine adenylyltransferase | | Authors: | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-12-26 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of the complex of trimeric Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with citrate ion at 1.87 A resolution
To Be Published
|
|
7C7M
 
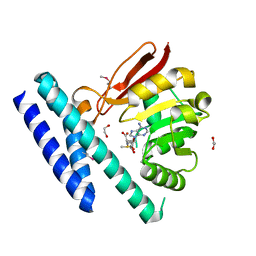 | | The structure of SAM-bound CntL, an aminobutyrate transferase in staphylopine biosysnthesis | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYLMETHIONINE, Staphylopine biosynthesis enzyme CntL | | Authors: | Luo, Z, Luo, S, Zhou, H. | | Deposit date: | 2020-05-26 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the ligand recognition and catalysis of the key aminobutanoyltransferase CntL in staphylopine biosynthesis.
Faseb J., 35, 2021
|
|
5L22
 
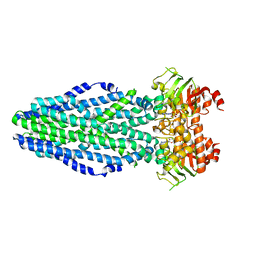 | | PrtD T1SS ABC transporter | | Descriptor: | ABC transporter (HlyB subfamily), ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Morgan, J.L.W, Zimmer, J. | | Deposit date: | 2016-07-30 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of a Type-1 Secretion System ABC Transporter.
Structure, 25, 2017
|
|
3GSJ
 
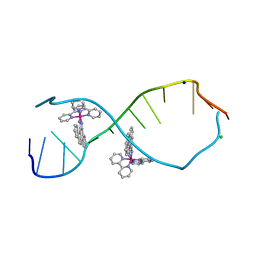 | | A Bulky Rhodium Complex Bound to an Adenosine-Adenosine DNA Mismatch | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*AP*TP*TP*AP*CP*CP*G)-3', CHLORIDE ION, SODIUM ION, ... | | Authors: | Zeglis, B.M, Pierre, V.C, Kaiser, J.T, Barton, J.K. | | Deposit date: | 2009-03-27 | | Release date: | 2009-05-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A bulky rhodium complex bound to an adenosine-adenosine DNA mismatch: general architecture of the metalloinsertion binding mode
Biochemistry, 48, 2009
|
|
7XEB
 
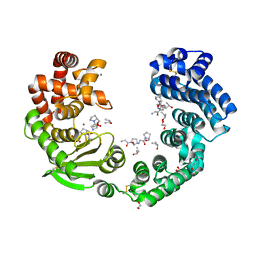 | | Collagenase from Grimontia (Vibrio) hollisae 1706B complexed with Gly-Pro-Hyp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLY-PRO-HYP peptide, ... | | Authors: | Ikeuchi, T, Yasumoto, M, Takita, T, Mizutani, K, Mikami, B, Tanaka, K, Hattori, S, Yasukawa, K. | | Deposit date: | 2022-03-30 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of Grimontia hollisae collagenase provides insights into its novel substrate specificity toward collagen.
J.Biol.Chem., 298, 2022
|
|
5ZI9
 
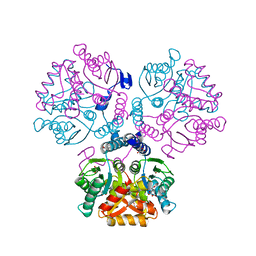 | | Crystal structure of type-II LOG from Streptomyces coelicolor A3 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Cytokinin riboside 5'-monophosphate phosphoribohydrolase, ... | | Authors: | Seo, H, Kim, K.-J. | | Deposit date: | 2018-03-14 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical characterization of the type-II LOG protein from Streptomyces coelicolor A3.
Biochem. Biophys. Res. Commun., 499, 2018
|
|
6RN6
 
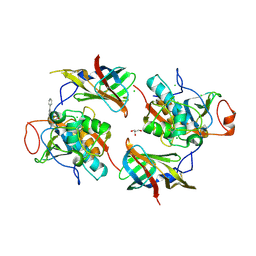 | | DPP1 in complex with inhibitor | | Descriptor: | (2~{S},4~{R})-~{N}-[(2~{S})-1-azanyl-3-(4-phenylphenyl)propan-2-yl]-4-oxidanyl-pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kack, H. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DPP1 Inhibitors: Exploring the Role of Water in the S2 Pocket of DPP1 with Substituted Pyrrolidines.
Acs Med.Chem.Lett., 10, 2019
|
|
5YUM
 
 | | Crystallographic structures of IlvN.Val/Ile complexes:Conformational selectivity for feedback inhibition of AHASs | | Descriptor: | Acetolactate synthase isozyme 1 small subunit, COBALT (II) ION, ISOLEUCINE | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-22 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.432 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
