5XXG
 
 | |
8RHZ
 
 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2023-12-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4W9F
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 5) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | van Molle, I, Hewitt, S, Galdeano, C, Gadd, M.S, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
9N69
 
 | | Structure of the retron IA complex with HNH nuclease in the "down" orientation | | Descriptor: | AAA family ATPase, ADENOSINE-5'-TRIPHOSPHATE, RNA-directed DNA polymerase, ... | | Authors: | Burman, N, Thomas-George, J, Wilkinson, R, Wiedenheft, B. | | Deposit date: | 2025-02-05 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis of antiphage defense by an ATPase-associated reverse transcriptase.
Biorxiv, 2025
|
|
6TTU
 
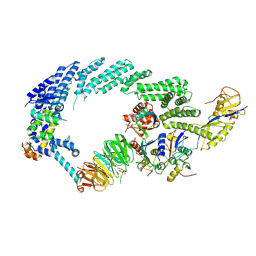 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | Descriptor: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
7QY7
 
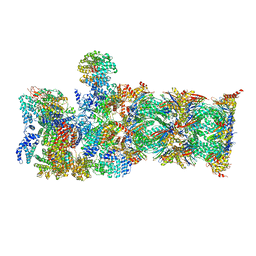 | | Proteasome-ZFAND5 Complex Z-A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXP
 
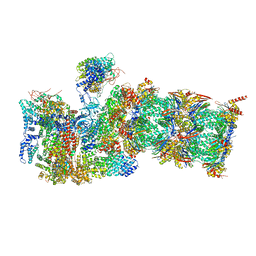 | | Proteasome-ZFAND5 Complex Z+B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXU
 
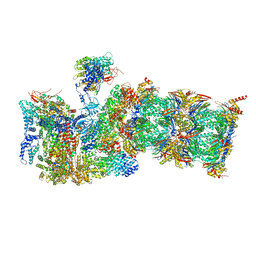 | | Proteasome-ZFAND5 Complex Z+C state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXW
 
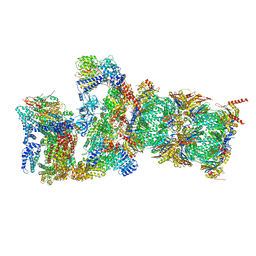 | | Proteasome-ZFAND5 Complex Z+D state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYA
 
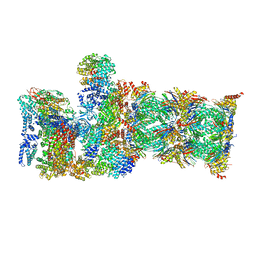 | | Proteasome-ZFAND5 Complex Z-B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXN
 
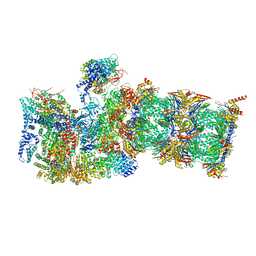 | | Proteasome-ZFAND5 Complex Z+A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYB
 
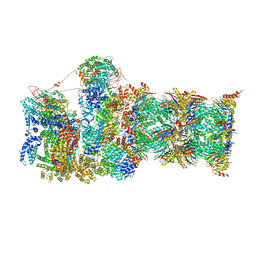 | | Proteasome-ZFAND5 Complex Z-C state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
2I5W
 
 | |
7QXX
 
 | | Proteasome-ZFAND5 Complex Z+E state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
9N6C
 
 | | Structure of the Retron IA Complex without the HNH Nuclease | | Descriptor: | AAA family ATPase, ADENOSINE-5'-TRIPHOSPHATE, RNA-directed DNA polymerase, ... | | Authors: | Burman, N, Thomas-George, J, Wilkinson, R, Wiedenheft, B. | | Deposit date: | 2025-02-05 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural basis of antiphage defense by an ATPase-associated reverse transcriptase.
Biorxiv, 2025
|
|
4LOO
 
 | | Structural basis of autoactivation of p38 alpha induced by TAB1 (Monoclinic crystal form) | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, Mitogen-activated protein kinase 14, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Chaikuad, A, DeNicola, G.F, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Marber, M.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism and consequence of the autoactivation of p38 alpha mitogen-activated protein kinase promoted by TAB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4LOQ
 
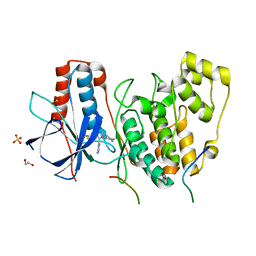 | | Structural basis of autoactivation of p38 alpha induced by TAB1 (Tetragonal crystal form with bound sulphate) | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, Mitogen-activated protein kinase 14, ... | | Authors: | Chaikuad, A, DeNicola, G.F, Yue, W.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Marber, M.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Mechanism and consequence of the autoactivation of p38 alpha mitogen-activated protein kinase promoted by TAB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
9O4L
 
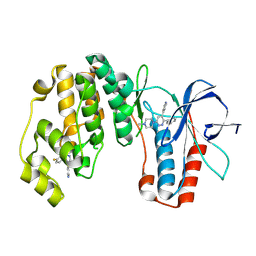 | |
9NYT
 
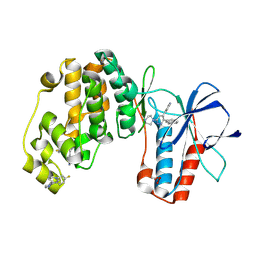 | | Crystal structure of Human p38 alpha MAPK in Complex with MW01-32-154JS | | Descriptor: | (3P)-3-(naphthalen-2-yl)-6-(piperazin-1-yl)-4-(pyridin-4-yl)pyridazine, 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, ETHANEPEROXOIC ACID, ... | | Authors: | Brunzelle, J.S, Shuvalova, L, Roy, S.M, Watterson, D.M. | | Deposit date: | 2025-03-28 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Human p38 alpha MAPK in Complex with
MW01-32-154JS
To Be Published
|
|
9M0D
 
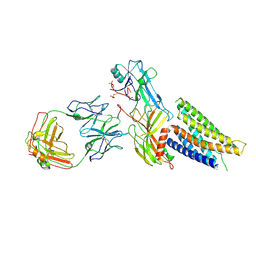 | | Cryo-EM structure of neurotensin receptor 1 in complex with beta-arrestin 1 | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Sun, D.M, Li, X, Yuan, Q.N, Tian, C.L. | | Deposit date: | 2025-02-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular mechanism of the arrestin-biased agonism of neurotensin receptor 1 by an intracellular allosteric modulator.
Cell Res., 35, 2025
|
|
5Y1U
 
 | | Crystal structure of RBBP4 bound to AEBP2 RRK motif | | Descriptor: | Histone-binding protein RBBP4, SULFATE ION, Zinc finger protein AEBP2 | | Authors: | Sun, A, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2017-07-21 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4
Protein Cell, 2017
|
|
6LTH
 
 | | Structure of human BAF Base module | | Descriptor: | AT-rich interactive domain-containing protein 1A, SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1, ... | | Authors: | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
5XS2
 
 | |
1KXU
 
 | | CYCLIN H, A POSITIVE REGULATORY SUBUNIT OF CDK ACTIVATING KINASE | | Descriptor: | CYCLIN H | | Authors: | Kim, K.K, Chamberin, H.M, Morgan, D.O, Kim, S.-H. | | Deposit date: | 1996-08-08 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of human cyclin H, a positive regulator of the CDK-activating kinase.
Nat.Struct.Biol., 3, 1996
|
|
8AEL
 
 | | SYNJ2BP complex with a synthetic Vangl2 peptide (3mer). | | Descriptor: | CALCIUM ION, GLY-GLY-GLY-THR-SER-VAL, GLYCEROL, ... | | Authors: | Carrasco, K, Cousido Siah, A, Gogl, G, Betzi, S, McEwen, A, Kostmann, C, Trave, G. | | Deposit date: | 2022-07-13 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SYNJ2BP PDZ domain in complex with a synthetic Vangl2 peptide.
To Be Published
|
|
