6FCK
 
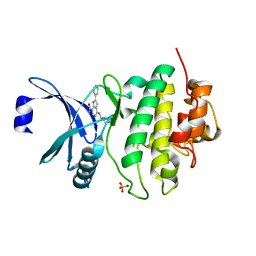 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 13 | | Descriptor: | 2-phenyl-4-[[(3~{S})-piperidin-3-yl]amino]-1~{H}-indole-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
3FXA
 
 | |
9EKH
 
 | | Cryo-EM structure ONO3030297-bound prostaglandin D2 receptor (DP1)-bRIL-Fab complex | | Descriptor: | 1-[2-chloro-5-(2,6-dimethyl-4-{[(2S)-4-methyl-3,4-dihydro-2H-1,4-benzoxazin-2-yl]methoxy}benzamido)phenyl]cyclopropane-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Prostaglandin D2 receptor,Soluble cytochrome b562, ... | | Authors: | Davoudinasab, B, Cherezov, V, Han, G.W, Kim, D. | | Deposit date: | 2024-12-02 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into the mechanism of activation and inhibition of the prostaglandin D2 receptor 1.
Nat Commun, 16, 2025
|
|
7YKO
 
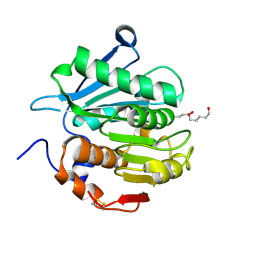 | | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol | | Descriptor: | Triacylglycerol lipase, pentane-1,5-diol | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol
To Be Published
|
|
6D2P
 
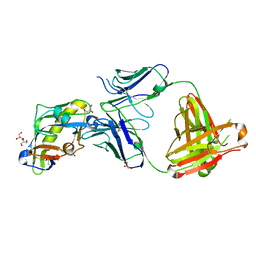 | |
6CJB
 
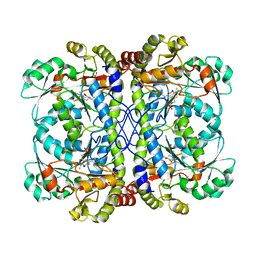 | |
4FS7
 
 | |
5BXG
 
 | |
6CYV
 
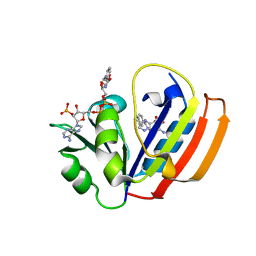 | | E. coli DHFR ternary complex with NADP and dihydrofolate | | Descriptor: | DIHYDROFOLIC ACID, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6AWA
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
5BMT
 
 | |
6ALV
 
 | | Crystal structure of H107A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant (no CuH bound) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
4XS2
 
 | | Irak4-inhibitor co-structure | | Descriptor: | (1R,2S,3R,5R)-3-({5-(1,3-benzothiazol-2-yl)-6-chloro-2-[(3-methoxypropyl)amino]pyrimidin-4-yl}amino)-5-(hydroxymethyl)cyclopentane-1,2-diol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-01-21 | | Release date: | 2015-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery and hit-to-lead optimization of 2,6-diaminopyrimidine inhibitors of interleukin-1 receptor-associated kinase 4.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
9S04
 
 | | PYCR1 in complex with 1-(2,4-Difluorophenyl)-2-(1H-1,2,4-triazol-1- yl)ethanone | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,4-bis(fluoranyl)phenyl]-2-(1,2,4-triazol-1-yl)ethanone, Isoform 3 of Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Ragin-Oh, W, Czerwonka, D, Ruszkowski, M. | | Deposit date: | 2025-07-16 | | Release date: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic fragment screening reveals new starting points for PYCR1 inhibitor design.
Bioorg.Chem., 165, 2025
|
|
9PQ6
 
 | | MBP-Mcl1 in complex with ligand 12 | | Descriptor: | 1,2-ETHANEDIOL, 17-chloranyl-33-fluoranyl-5,13,14,22-tetramethyl-28-oxa-9-thia-5,6,12,13,24-pentazaheptacyclo[27.7.1.1^{4,7}.0^{11,15}.0^{16,21}.0^{20,24}.0^{30,35}]octatriaconta-1(36),4(38),6,11,14,16,18,20,22,29(37),30(35),31,33-tridecaene-23-carboxylic acid, FORMIC ACID, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2025-07-22 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | In Pursuit of Best-in-Class MCL-1 Inhibitors: Discovery of Highly Potent 1,4-Indoyl Macrocycles with Favorable Physicochemical Properties.
J.Med.Chem., 68, 2025
|
|
7KI7
 
 | |
3T6K
 
 | |
6CZY
 
 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with Pyridoxamine-5'-phosphate (PMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
9PQ7
 
 | | MBP-Mcl1 in complex with ligand 21b | | Descriptor: | 1,2-ETHANEDIOL, 17-chloranyl-33-fluoranyl-12-[2-(2-methoxyethoxy)ethyl]-5,14,22-trimethyl-28-oxa-9-thia-5,6,12,13,24-pentazaheptacyclo[27.7.1.1^{4,7}.0^{11,15}.0^{16,21}.0^{20,24}.0^{30,35}]octatriaconta-1(36),4(38),6,11(15),13,16,18,20,22,29(37),30(35),31,33-tridecaene-23-carboxylic acid, FORMIC ACID, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2025-07-22 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | In Pursuit of Best-in-Class MCL-1 Inhibitors: Discovery of Highly Potent 1,4-Indoyl Macrocycles with Favorable Physicochemical Properties.
J.Med.Chem., 68, 2025
|
|
9N0V
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, KFA-027 | | Descriptor: | Capsid protein p24, N-[(1S)-1-{(3M)-3-[4-chloro-3-(methanesulfonamido)-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl]-6-[3-(methanesulfonyl)-3-methylbut-1-yn-1-yl]pyridin-2-yl}-2-phenylethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Dinh, T.T, Kvaratskhelia, M. | | Deposit date: | 2025-01-24 | | Release date: | 2025-06-18 | | Last modified: | 2025-11-05 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Delineating Structural Functionalities of Lenacapavir Amenable to Modifications for Targeting Emerging Drug-Resistant HIV‐1 Capsid Variants.
Acs Med.Chem.Lett., 16, 2025
|
|
7ADV
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ447 (compound 6v) | | Descriptor: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[2-(2-morpholin-4-ylethylsulfonyl)ethyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Pye, V.E, Cherepanov, P. | | Deposit date: | 2020-09-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | HIV-1 Integrase Inhibitors with Modifications That Affect Their Potencies against Drug Resistant Integrase Mutants.
Acs Infect Dis., 7, 2021
|
|
7ADU
 
 | |
6CJY
 
 | | Crystal Structure of Mnk2-D228G in complex with Inhibitor | | Descriptor: | 5-[(7H-purin-6-yl)amino]-1H-isoindol-1-one, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CK3
 
 | | Co-crytsal Structure of MNK2 in Complex With an Inhibitor | | Descriptor: | (3R)-3-methyl-5-[(pyrimidin-4-yl)amino]-2,3-dihydro-1H-isoindol-1-one, CHLORIDE ION, MAP kinase-interacting serine/threonine-protein kinase 2, ... | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
5ETE
 
 | |
