6FX4
 
 | |
3ODZ
 
 | | Crystal structure of P38alpha Y323R active mutant | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
6FV6
 
 | | Monomer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Monomer structure of Aq128 in the outward-facing state
To Be Published
|
|
6FV8
 
 | | Dimer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dimer structure of the MATE family multidrug resistance transporter Aq128
in the outward-facing state
To Be Published
|
|
3OEF
 
 | | Crystal structure of Y323F inactive mutant of p38alpha MAP kinase | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
2WU4
 
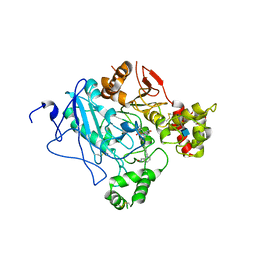 | | CRYSTAL STRUCTURE OF MOUSE ACETYLCHOLINESTERASE IN COMPLEX WITH FENAMIPHOS AND ORTHO-7 | | Descriptor: | 1,7-HEPTYLENE-BIS-N,N'-SYN-2-PYRIDINIUMALDOXIME, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Hornberg, A, Artursson, E, Warme, R, Pang, Y.-P, Ekstrom, F. | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Oxime-Bound Fenamiphos-Acetylcholinesterases: Reactivation Involving Flipping of the His447 Ring to Form a Reactive Glu334-His447-Oxime Triad.
Biochem.Pharm., 79, 2010
|
|
2XG3
 
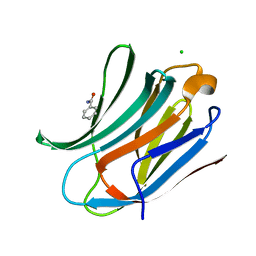 | | Human galectin-3 in complex with a benzamido-N-acetyllactoseamine inhibitor | | Descriptor: | BENZAMIDE, CHLORIDE ION, Galectin-3, ... | | Authors: | Diehl, C, Engstrom, O, Delaine, T, Hakansson, M, Genheden, S, Modig, K, Leffler, H, Ryde, U, Nilsson, U, Akke, M. | | Deposit date: | 2010-05-30 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3.
J. Am. Chem. Soc., 132, 2010
|
|
2XCI
 
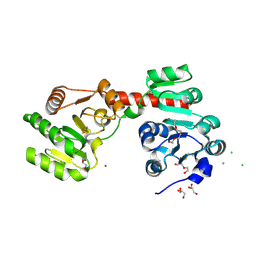 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6GHF
 
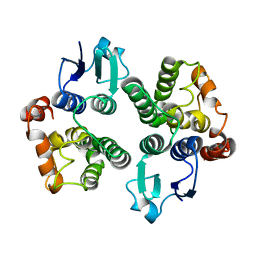 | | Crystal structure of a GST variant | | Descriptor: | PvGmGSTUG | | Authors: | Papageorgiou, A.C, Chronopoulou, E.G, Labrou, N.E. | | Deposit date: | 2018-05-07 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Expanding the Plant GSTome Through Directed Evolution: DNA Shuffling for the Generation of New Synthetic Enzymes With Engineered Catalytic and Binding Properties.
Front Plant Sci, 9, 2018
|
|
6EN4
 
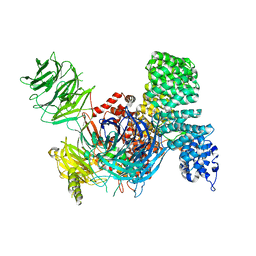 | | SF3b core in complex with a splicing modulator | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, Splicing factor 3B subunit 3, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2017-10-04 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural Basis of Splicing Modulation by Antitumor Macrolide Compounds.
Mol. Cell, 70, 2018
|
|
2XYC
 
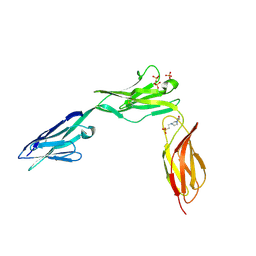 | | CRYSTAL STRUCTURE OF NCAM2 IGIV-FN3I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NEURAL CELL ADHESION MOLECULE 2, ... | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2XN2
 
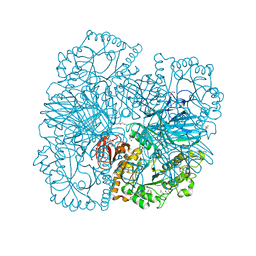 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
6GGU
 
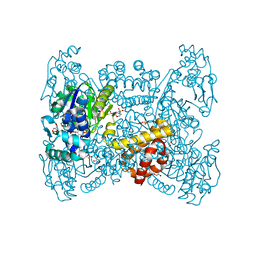 | | Crystal structure of native FE-hydrogenase from Methanothermobacter marburgensis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5,10-methenyltetrahydromethanopterin hydrogenase, GLYCEROL, ... | | Authors: | Wagner, T, Huang, G, Ermler, U, Shima, S. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Self-protection of the catalytic iron center of a methanogenic [Fe]-hydrogenase via a dynamic dimer-to-hexamer transformation
To Be Published
|
|
6GWH
 
 | |
3N5M
 
 | | Crystals structure of a Bacillus anthracis aminotransferase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, CHLORIDE ION, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, DiLeo, R, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystals structure of a Bacillus anthracis aminotransferase
TO BE PUBLISHED
|
|
6GF1
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | SULFATE ION, Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
3PZS
 
 | | Crystal Structure of a pyridoxamine kinase from Yersinia pestis CO92 | | Descriptor: | BETA-MERCAPTOETHANOL, Pyridoxamine kinase, SODIUM ION, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Kudritska, M, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-14 | | Release date: | 2011-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of a pyridoxamine kinase from Yersinia pestis CO92
TO BE PUBLISHED
|
|
2XY2
 
 | | CRYSTAL STRUCTURE OF NCAM2 IG1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2Y2V
 
 | | Nonaged form of Mouse Acetylcholinesterase inhibited by sarin-Update | | Descriptor: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-ETHOXYETHANOL, ... | | Authors: | Akfur, C, Artursson, E, Ekstrom, F. | | Deposit date: | 2010-12-16 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Methylphosphonate Adducts of Acetylcholinesterase Investigated by Time Correlated Single Photon Counting and X-Ray Crystallography
To be Published
|
|
2Y5E
 
 | | BARLEY LIMIT DEXTRINASE IN COMPLEX WITH ALPHA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Vester-Christensen, M.B, Hachem, M.A, Svensson, B, Henriksen, A. | | Deposit date: | 2011-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal Structure of an Essential Enzyme in Seed Starch Degradation: Barley Limit Dextrinase in Complex with Cyclodextrins.
J.Mol.Biol., 403, 2010
|
|
2Y37
 
 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 2-[(1R)-3-amino-1-phenyl-propoxy]-4-chloro-benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T.N, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2010-12-19 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y2U
 
 | | Nonaged form of Mouse Acetylcholinesterase inhibited by VX-Update | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ACETYLCHOLINESTERASE, ... | | Authors: | Akfur, C, Artursson, E, Ekstrom, F. | | Deposit date: | 2010-12-16 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Methylphosphonate Adducts of Acetylcholinesterase Investigated by Time Correlated Single Photon Counting and X-Ray Crystallography
To be Published
|
|
3Q58
 
 | | Structure of N-acetylmannosamine-6-Phosphate Epimerase from Salmonella enterica | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-27 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: |
|
|
2XF1
 
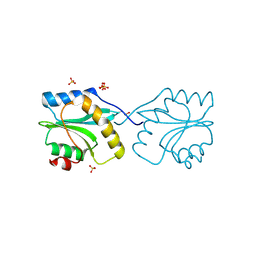 | | Crystal structure of Plasmodium falciparum actin depolymerization factor 1 | | Descriptor: | COFILIN ACTIN-DEPOLYMERIZING FACTOR HOMOLOG 1, SULFATE ION | | Authors: | Singh, B.K, Sattler, J.M, Huttu, J, Chatterjee, M, Schueler, H, Kursula, I. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures Explain Functional Differences in the Two Actin Depolymerization Factors of the Malaria Parasite.
J.Biol.Chem., 286, 2011
|
|
2XN1
 
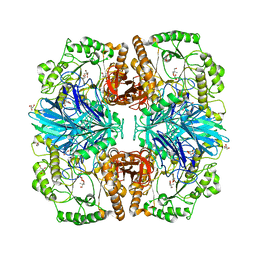 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-GALACTOSIDASE, GLYCEROL | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
