8EL1
 
 | | Structure of MBP-Mcl-1 in complex with ABBV-467 | | Descriptor: | (7R,16R)-19,23-dichloro-10-{[2-(4-{[(2R)-1,4-dioxan-2-yl]methoxy}phenyl)pyrimidin-4-yl]methoxy}-1-(4-fluorophenyl)-20,22-dimethyl-16-[(4-methylpiperazin-1-yl)methyl]-7,8,15,16-tetrahydro-18,21-etheno-13,9-(metheno)-6,14,17-trioxa-2-thia-3,5-diazacyclononadeca[1,2,3-cd]indene-7-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
8W28
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament bound to compound, XAV (focused refinement map). | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, Maltose/maltodextrin-binding periplasmic protein,Poly [ADP-ribose] polymerase tankyrase-2, ZINC ION | | Authors: | Malone, B.F, Zimmerman, J.L, Dow, L.E, Hite, R.K. | | Deposit date: | 2024-02-20 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | A potent and selective TNKS2 inhibitor for tumor-selective WNT suppression.
Biorxiv, 2025
|
|
8W23
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament bound to compound, TDI-2804 (consensus map). | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Poly [ADP-ribose] polymerase tankyrase-2, N-{2-[4-(2-hydroxypropan-2-yl)phenyl]-4-oxo-1,4-dihydroquinazolin-7-yl}-4-methoxy-6-phenylpyridine-3-carboxamide, ZINC ION | | Authors: | Malone, B.F, Zimmerman, J.L, Dow, L.E, Hite, R.K. | | Deposit date: | 2024-02-19 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | A potent and selective TNKS2 inhibitor for tumor-selective WNT suppression.
Biorxiv, 2025
|
|
8W25
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament bound to compound, TDI-2804 (focused refinement map). | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Poly [ADP-ribose] polymerase tankyrase-2, N-{2-[4-(2-hydroxypropan-2-yl)phenyl]-4-oxo-1,4-dihydroquinazolin-7-yl}-4-methoxy-6-phenylpyridine-3-carboxamide, ZINC ION | | Authors: | Malone, B.F, Zimmerman, J.L, Dow, L.E, Hite, R.K. | | Deposit date: | 2024-02-20 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | A potent and selective TNKS2 inhibitor for tumor-selective WNT suppression.
Biorxiv, 2025
|
|
8W27
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament bound to compound, XAV (consensus map). | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, Maltose/maltodextrin-binding periplasmic protein,Poly [ADP-ribose] polymerase tankyrase-2, ZINC ION | | Authors: | Malone, B.F, Zimmerman, J.L, Dow, L.E, Hite, R.K. | | Deposit date: | 2024-02-20 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | A potent and selective TNKS2 inhibitor for tumor-selective WNT suppression.
Biorxiv, 2025
|
|
8EKX
 
 | | Structure of MBP-Mcl-1 in complex with MIK665 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy]phenyl]propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
6TZC
 
 | | Crystal Structure of African Swine Fever Virus A179L with the Autophagy Regulator Beclin | | Descriptor: | Apoptosis regulator Bcl-2 homolog, Beclin-1, Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Banjara, S, Kvansakul, M, Hinds, M.G. | | Deposit date: | 2019-08-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of African Swine Fever Virus A179L with the Autophagy Regulator Beclin.
Viruses, 11, 2019
|
|
6SMV
 
 | | Structure of HPV49 E6 protein in complex with MAML1 LxxLL motif | | Descriptor: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Mastermind-like protein 1, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Kostmann, C, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Cellular target recognition by HPV18 and HPV49 oncoproteins
To be published
|
|
6SIV
 
 | | Structure of HPV16 E6 oncoprotein in complex with mutant IRF3 LxxLL motif | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-12 | | Release date: | 2019-08-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Deciphering the molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
7T31
 
 | | X-ray Structure of Clostridiodies difficile PilW | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Putative pilin protein chimera | | Authors: | Ronish, L.A, Piepenbrink, K.H. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of extracellular DNA by type IV pili promotes biofilm formation by Clostridioides difficile.
J.Biol.Chem., 298, 2022
|
|
3HST
 
 | |
8FNE
 
 | | phiPA3 PhuN Tetramer, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2022-12-27 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
5WVM
 
 | | Crystal structure of baeS cocrystallized with 2 mM indole | | Descriptor: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | Authors: | Wang, W, Zhang, Y, Rang, T, Xu, D. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
5WWG
 
 | | Crystal structure of hnRNPA2B1 in complex with AAGGACUUGC | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*AP*GP*GP*AP*CP*UP*U)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
3IOT
 
 | |
5V6Y
 
 | |
7TT6
 
 | | BamABCDE bound to substrate EspP in the intermediate-open EspP state | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TSZ
 
 | | BamABCDE bound to substrate EspP class 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT3
 
 | | BamABCDE bound to substrate EspP class 5 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT4
 
 | | BamABCDE bound to substrate EspP class 6 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT1
 
 | | BamABCDE bound to substrate EspP class 4 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT5
 
 | | BamABCDE bound to substrate EspP in the open-sheet EspP state | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT2
 
 | | BamABCDE bound to substrate EspP class 3 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
6SWR
 
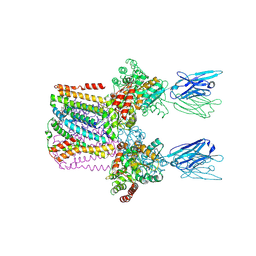 | | Crystal structure of the lysosomal potassium channel MtTMEM175 T38A mutant soaked with zinc | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody, Maltose/maltodextrin-binding periplasmic protein,Maltodextrin-binding protein,Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2019-09-23 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K + channels.
Elife, 9, 2020
|
|
7U0G
 
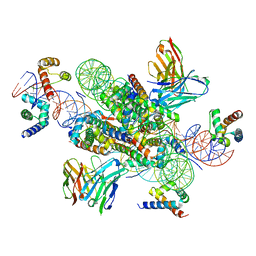 | | structure of LIN28b nucleosome bound 3 OCT4 | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
