7EJ0
 
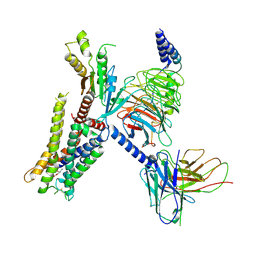 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex | | Descriptor: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJA
 
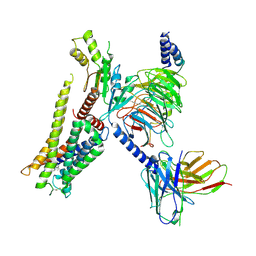 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to dexmedetomidine | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJ8
 
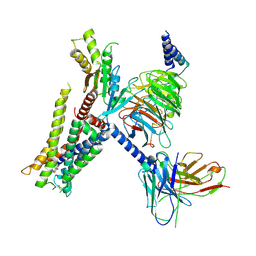 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to brimonidine | | Descriptor: | Alpha-2A adrenergic receptor, Brimonidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
4WQY
 
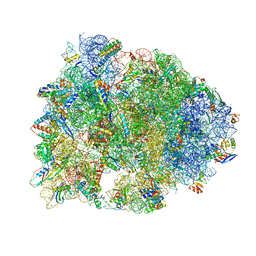 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the post-translocational state (without fusitic acid) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4WQU
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G trapped by the antibiotic dityromycin | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4WPO
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the pre-translocational state | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
4WQF
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G and fusidic acid in the post-translocational state | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4V6A
 
 | | Structure of EF-P bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-06-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome.
Science, 325, 2009
|
|
3JRQ
 
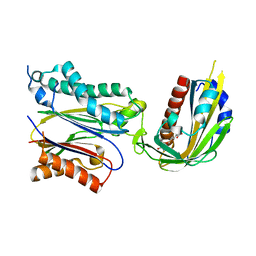 | | Crystal structure of (+)-ABA-bound PYL1 in complex with ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Protein phosphatase 2C 56, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
4V83
 
 | | Crystal structure of a complex containing domain 3 from the PSIV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
9KVL
 
 | | Neutron and X-ray joint refined structure of a copper-containing nitrite reductase (C135A mutant) in complex with nitrite | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Lintuluoto, M, Hirano, Y, Kusaka, K, Inoue, T, Tamada, T. | | Deposit date: | 2024-12-05 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-02 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Structural basis of cuproenzyme nitrite reduction at the level of a single hydrogen atom.
J.Biol.Chem., 301, 2025
|
|
4V55
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with gentamicin and ribosome recycling factor (RRF). | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V5K
 
 | | Structure of cytotoxic domain of colicin E3 bound to the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Meenan, N.A.G, Sharma, A, Kelley, A.C, Kleanthous, C, Ramakrishnan, V. | | Deposit date: | 2010-05-29 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for 16S Ribosomal RNA Cleavage by the Cytotoxic Domain of Colicin E3.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4V7Z
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with telithromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V8F
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-ttyr complex with paromomycin). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V8X
 
 | | Structure of Thermus thermophilus ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Feng, S, Chen, Y, Kamada, K, Wang, H, Tang, K, Wang, M, Gao, Y.G. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Yoeb-Ribosome Structure: A Canonical Rnase that Requires the Ribosome for its Specific Activity.
Nucleic Acids Res., 41, 2013
|
|
4WIZ
 
 | | Crystal structure of Grouper nervous necrosis virus-like particle at 3.6A | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
9MQ4
 
 | | Damaged 70S ribosome with PrfH bound | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Tian, Y, Li, Q, Jin, H, Fatma, S, Zeng, F, Huang, R.H. | | Deposit date: | 2025-01-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Molecular and structural basis of a subfamily of PrfH rescuing both the damaged and intact ribosomes stalled in translation.
Biorxiv, 2025
|
|
9MOR
 
 | | Damaged 70S ribosome with PrfH bound | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Tian, Y, Li, Q, Jin, H, Fatma, S, Zeng, F, Huang, R.H. | | Deposit date: | 2024-12-27 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Molecular and structural basis of a subfamily of PrfH rescuing both the damaged and intact ribosomes stalled in translation.
Biorxiv, 2025
|
|
3JC8
 
 | | Architectural model of the type IVa pilus machine in a piliated state | | Descriptor: | LysM domain protein, PilA, PilN, ... | | Authors: | Chang, Y.-W, Rettberg, L.A, Jensen, G.J. | | Deposit date: | 2015-11-24 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY | | Cite: | Architecture of the type IVa pilus machine.
Science, 351, 2016
|
|
6DZU
 
 | |
6BY1
 
 | | E. coli pH03H9 complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Amiri, H, Noller, H.F. | | Deposit date: | 2017-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Structural evidence for product stabilization by the ribosomal mRNA helicase.
Rna, 25, 2019
|
|
6E6J
 
 | | BRD2_Bromodomain2 complex with inhibitor 744 | | Descriptor: | Bromodomain-containing protein 2, N-ethyl-4-[2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl]-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Longenecker, K.L, Park, C.H, Bigelow, L. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.
Nature, 578, 2020
|
|
6E2R
 
 | | Mechanism of cellular recognition by PCV2 | | Descriptor: | Capsid protein of PCV2 | | Authors: | Khayat, R, Dhindwal, S. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid.
J.Virol., 93, 2019
|
|
