5KMB
 
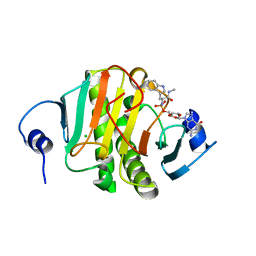 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside L-Trp phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~ {S})-3-(1~{H}-indol-3-yl)-1-(methylamino)-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
3DOL
 
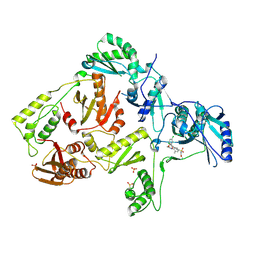 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with GW695634. | | Descriptor: | N-({4-[({4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}acetyl)amino]-3-methylphenyl}sulfonyl)propanamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
7LUY
 
 | |
5CCK
 
 | |
3FWP
 
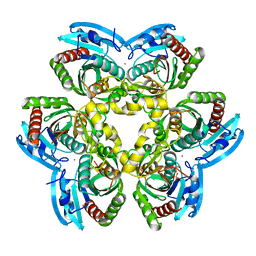 | | X-ray structure of uridine nucleoside phosphorylease from Salmonella typhimurium complexed with phosphate and its inhibitor 2,2'-anhydrouridine at 1.86 A resolution | | Descriptor: | 2,2'-Anhydro-(1-beta-D-arabinofuranosyl)uracil, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Lashkov, S.A, Mikhailov, A.M, Gabdulkhakov, A.G. | | Deposit date: | 2009-01-19 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The X-ray structure of Salmonella typhimurium uridine nucleoside phosphorylase complexed with 2,2'-anhydrouridine, phosphate and potassium ions at 1.86 A resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6TZ6
 
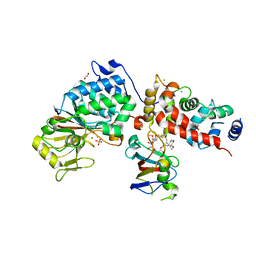 | | Crystal Structure of Candida Albicans Calcineurin A, Calcineurin B, FKBP12 and FK506 (Tacrolimus) | | Descriptor: | 1,2-ETHANEDIOL, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2019-08-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents.
Nat Commun, 10, 2019
|
|
6UKE
 
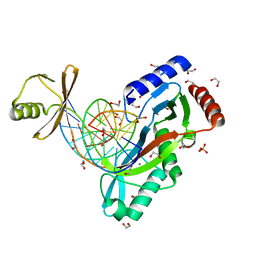 | |
9R0Q
 
 | | Paraoxonase-1 in complex with terbium(III) and 2-hydroxyquinoline | | Descriptor: | BROMIDE ION, CALCIUM ION, QUINOLIN-2(1H)-ONE, ... | | Authors: | Smerkolj, J, Pavsic, M, Golicnik, M. | | Deposit date: | 2025-04-24 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Intramolecular sensitization and structure of a Tb 3+ /2-hydroxyquinoline conjugate in the paraoxonase 1 active site.
Dalton Trans, 54, 2025
|
|
5W0F
 
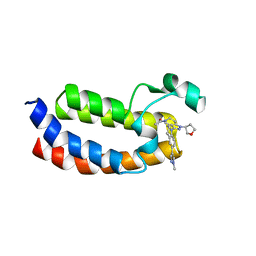 | | CREBBP Bromodomain in complex with Cpd3 ((S)-1-(3-(6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
5W0L
 
 | | CREBBP Bromodomain in complex with Cpd10 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-31 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
9WP9
 
 | | Cryo-EM structure of the d18:1 S1P-bound S1PR3 and Gq complex | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Engineered G-alpha-q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Im, D, Asada, H, Iwata, S, Yamauchi, M, Hagiwara, M. | | Deposit date: | 2025-09-08 | | Release date: | 2025-11-26 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into the G-protein subtype selectivity revealed by human sphingosine-1-phosphate receptor 3-G q complexes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9PGU
 
 | | HIV Capsid Hexamer bound to Compound 40 | | Descriptor: | (5M)-5-{2-[(1S)-2-(3,5-difluorophenyl)-1-{2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamido}ethyl]pyridin-3-yl}-2-fluorobenzamide, HIV-1 capsid | | Authors: | Somoza, J.R, Anderson, R.L, Villasenor, A.G, Ferrao, R.D. | | Deposit date: | 2025-07-08 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Discovery of Lenacapavir: First-in-Class Twice-Yearly Capsid Inhibitor for HIV-1 Treatment and Pre-exposure Prophylaxis.
J.Med.Chem., 68, 2025
|
|
9NPH
 
 | | X-ray crystal structure of recombinant Can f 1 in complex with human IgE mAb 1J11 Fab | | Descriptor: | 1J11 Fab heavy chain, 1J11 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Khatri, K, Ball, A, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2025-03-11 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Human IgE monoclonal antibodies define two unusual epitopes trapping dog allergen Can f 1 in different conformations.
Protein Sci., 34, 2025
|
|
9Q9N
 
 | | HSV-1 prefusion glycoprotein B bound by Nb1_gbHSV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein B, Nb1_gbHSV | | Authors: | Vollmer, B, Mulvaney, T, Ebel, H, Nentwig, J, Gruenewald, K. | | Deposit date: | 2025-02-26 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A nanobody specific to prefusion glycoprotein B neutralizes HSV-1 and HSV-2.
Nature, 646, 2025
|
|
9PQ9
 
 | | Fem-1 homolog B (FEM1B) in complex with VU0421763 | | Descriptor: | 6-(propylsulfanyl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, Protein fem-1 homolog B, SULFATE ION | | Authors: | Katinas, J.M, Fesik, S.W. | | Deposit date: | 2025-07-22 | | Release date: | 2025-11-26 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Nuclear Magnetic Resonance-based fragment screen of the E3 ligase Fem-1 homolog B.
Protein Sci., 34, 2025
|
|
9R8B
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE PLK1 KINASE DOMAIN IN COMPLEX WITH THIAZOLIDINONE INHIBITOR COMPOUND 1 AND A SELECTIVE DARPIN | | Descriptor: | (2~{Z})-2-cyano-2-[3-ethyl-5-[(~{E})-[2-[methyl-(1-methylpiperidin-4-yl)amino]pyridin-4-yl]iminomethyl]-4-oxidanylidene-1,3-thiazol-2-ylidene]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, CHLORIDE ION, DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN), ... | | Authors: | Hillig, R.C, Matias, P.M, Bandeiras, T.M, Siemeister, G, Schulze, V.K, Schmitz, A.A, Eberspaecher, U. | | Deposit date: | 2025-05-16 | | Release date: | 2025-11-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Polo-like kinase 1–inhibitor co-complex structures via the surface-entropy reduction approach and a DARPin-assisted approach
Acta Crystallogr.,Sect.D, 81, 2025
|
|
9PQA
 
 | |
9PXO
 
 | | Fem-1 homolog B (FEM1B) in complex with VU0023775 | | Descriptor: | 2-tert-butyl-1,3-dioxo-2,3-dihydro-1H-isoindole-5-carboxylic acid, Protein fem-1 homolog B | | Authors: | Katinas, J.M, Fesik, S.W. | | Deposit date: | 2025-08-06 | | Release date: | 2025-11-26 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Nuclear Magnetic Resonance-based fragment screen of the E3 ligase Fem-1 homolog B.
Protein Sci., 34, 2025
|
|
9S6W
 
 | | HIV-1 capsid (M-group) - native in complex with JW3-100 | | Descriptor: | (S)-N-(1-(5-((2-amino-2-oxoethyl)thio)-4-(4-(tert-Butyl)phenyl)-4H-1,2,4-triazol-3-yl)-2-(3,5-difluorophenyl)ethyl)-2-(5-hydroxy-1H-indol-3-yl)acetamide, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Govasli, M.A.L, Pinotsis, N, Towers, G, Selwood, D, Jacques, D.A. | | Deposit date: | 2025-08-01 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | Cofactor-mimicking HIV-1 capsid inhibitors, and their escape mutants, drive innate immune sensing
To Be Published
|
|
9PWJ
 
 | |
5IEE
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with 1-deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9U9C
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed amoxicillin | | Descriptor: | (2~{R},4~{S})-2-[(1~{R})-1-[[(2~{R})-2-azanyl-2-(4-hydroxyphenyl)ethanoyl]amino]-2-oxidanyl-2-oxidanylidene-ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Shi, X, Zhang, Q, Liu, W. | | Deposit date: | 2025-03-27 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure reveals the hydrophilic R1 group impairs NDM-1-ligand binding via water penetration at L3.
J Struct Biol X, 12, 2025
|
|
9PW6
 
 | | Myeloid cell leukemia-1 (Mcl-1) complexed with compound 8 | | Descriptor: | 7-[(4R,5S,6P)-7-chloro-10-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-4-methyl-1-oxo-6-(1,3,5-trimethyl-1H-pyrazol-4-yl)-3,4-dihydropyrazino[1,2-a]indol-2(1H)-yl]-1-(2-methoxyethyl)-5-methyl-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B, Fesik, S.W. | | Deposit date: | 2025-08-04 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Discovery of Macrocyclic Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors that Demonstrate Potent Cellular Efficacy and In Vivo Activity in a Mouse Solid Tumor Xenograft Model.
J.Med.Chem., 68, 2025
|
|
6RQK
 
 | | Crystal structure of GH125 1,6-alpha-mannosidase from Clostridium perfringens in complex with mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Alpha-1,6-mannosidase | | Authors: | Males, A, Davies, G.J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distortion of mannoimidazole supports a B2,5boat transition state for the family GH125 alpha-1,6-mannosidase from Clostridium perfringens.
Org.Biomol.Chem., 17, 2019
|
|
5EHO
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, ~{N}8-cyclohexyl-~{N}2-[2-methoxy-4-(1-methylpyrazol-4-yl)phenyl]pyrido[3,4-d]pyrimidine-2,8-diamine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach
To Be Published
|
|
