6X9U
 
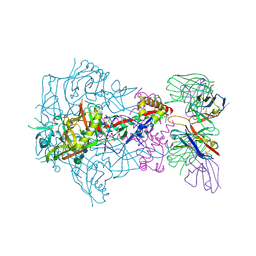 | | HIV-1 Envelope Glycoprotein BG505 SOSIP.664, expressed in HEK293S cells and partially deglycosylated by endoglycosidase H, in complex with RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp120, ... | | Authors: | Berndsen, Z.T, Ward, A.B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualization of the HIV-1 Env glycan shield across scales.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8BOY
 
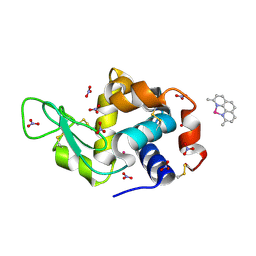 | | X-ray structure of the adduct formed upon reaction of the five-coordinate Pt(II) complex, 1-Me,Me, with HEWL at pH 4.0 | | Descriptor: | 1-[1,3-dimethyl-4-(1~{H}-1,2,3-triazol-5-yl)imidazol-1-ium-2-yl]-1,2',11'-trimethyl-spiro[1$l^{6}-platinacycloprop-2-ene-1,15'-1,12-diaza-15$l^{6}-platinatetracyclo[10.2.1.0^{5,14}.0^{8,13}]pentadeca-2,4,6,8,10,13-hexaene], Lysozyme C, NITRATE ION, ... | | Authors: | Ferraro, G, Tito, G, Merlino, A. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Impact of Hydrophobic Chains in Five-Coordinate Glucoconjugate Pt(II) Anticancer Agents.
Int J Mol Sci, 24, 2023
|
|
6R4I
 
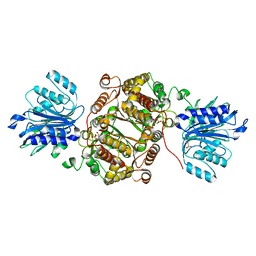 | | Crystal structure of human GFAT-1 G461E | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6X9V
 
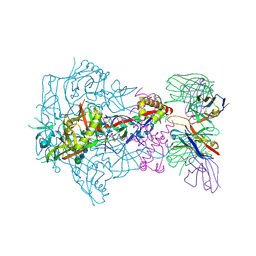 | | HIV-1 Envelope Glycoprotein BG505 SOSIP.664, expressed in HEK293S cells and deglycosylated by endoglycosidase H, in complex with RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp120, ... | | Authors: | Berndsen, Z.T, Ward, A.B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualization of the HIV-1 Env glycan shield across scales.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7N8K
 
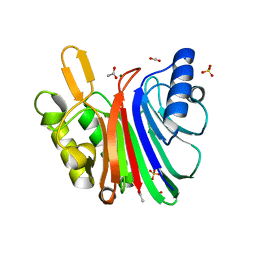 | | LINE-1 endonuclease domain complex with Mg | | Descriptor: | ACETATE ION, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | Korolev, S, Miller, I. | | Deposit date: | 2021-06-15 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural dissection of sequence recognition and catalytic mechanism of human LINE-1 endonuclease.
Nucleic Acids Res., 49, 2021
|
|
7NB4
 
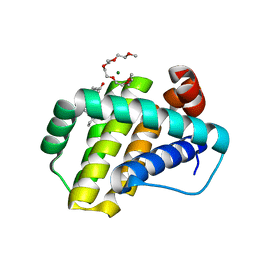 | | Structure of Mcl-1 complex with compound 1 | | Descriptor: | (2~{R})-2-[[5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
9HDO
 
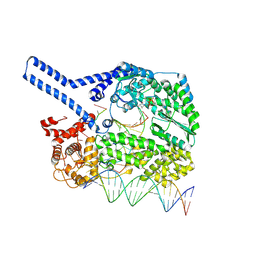 | | The Human LINE-1 ORF2p target-primed reverse transcription complex | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | Ghanim, G.E, Hu, H, Nguyen, T.H.D. | | Deposit date: | 2024-11-12 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural mechanism of LINE-1 target-primed reverse transcription.
Science, 388, 2025
|
|
9HDR
 
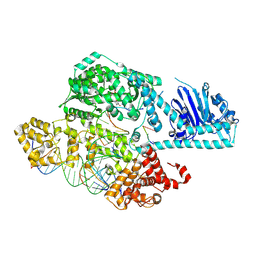 | | Human LINE-1 ORF2p target-primed reverse transcription complex with EN domain resolved | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | Ghanim, G.E, Hu, H, Nguyen, T.H.D. | | Deposit date: | 2024-11-12 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanism of LINE-1 target-primed reverse transcription.
Science, 388, 2025
|
|
9HDQ
 
 | | The Human LINE-1 ORF2p target-primed reverse transcription complex with the fingers domain in an open conformation | | Descriptor: | LINE-1 retrotransposable element ORF2 protein, Target DNA strand 1, Target DNA strand 2, ... | | Authors: | Ghanim, G.E, Hu, H, Nguyen, T.H.D. | | Deposit date: | 2024-11-12 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural mechanism of LINE-1 target-primed reverse transcription.
Science, 388, 2025
|
|
6FMF
 
 | | Neuropilin-1 b1 domain in complex with EG01377; 2.8 Angstrom structure | | Descriptor: | (2~{S})-2-[[3-[[5-[4-(aminomethyl)phenyl]-1-benzofuran-7-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1, trifluoroacetic acid | | Authors: | Yelland, T, Djordjevic, S, Selwood, D, Zachary, I, Frankel, P. | | Deposit date: | 2018-01-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta ) Production in Regulatory T-Cells.
J. Med. Chem., 61, 2018
|
|
5K0I
 
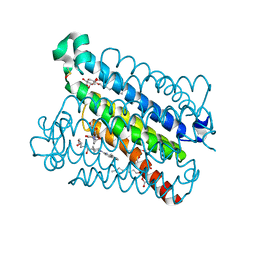 | | mpges1 bound to an inhibitor | | Descriptor: | 1,5-anhydro-2,3,4-trideoxy-3-{[(4S)-3,3-dimethyl-1-(8-methylquinolin-2-yl)piperidine-4-carbonyl]amino}-D-erythro-hexitol, GLUTATHIONE, Prostaglandin E synthase, ... | | Authors: | Luz, J.G, Kuklish, S.L. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-14 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of 3,3-dimethyl substituted N-aryl piperidines as potent microsomal prostaglandin E synthase-1 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
9HDP
 
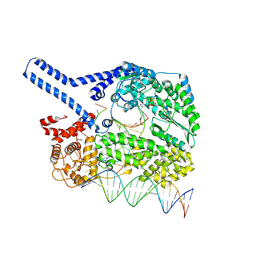 | | The Human LINE-1 ORF2p target-primed reverse transcription complex with the fingers domain in a closed conformation | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | Ghanim, G.E, Hu, H, Nguyen, T.H.D. | | Deposit date: | 2024-11-12 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural mechanism of LINE-1 target-primed reverse transcription.
Science, 388, 2025
|
|
3CNY
 
 | |
7L4T
 
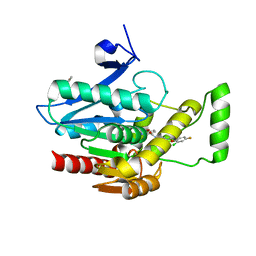 | | Crystal structure of human monoacylglycerol lipase in complex with compound 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidine-1-carbonyl}-2H-1,4-benzoxazin-3(4H)-one, ACETATE ION, ... | | Authors: | Qin, L, Gay, S.C, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7C7X
 
 | |
5L0U
 
 | | human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EGF(+), ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
9MTW
 
 | | Crystal structure of ADC-1-ertapenem complex | | Descriptor: | (2S,3R,4R)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, ... | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2025-01-12 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of carbapenemase activity in the class C beta-lactamase ADC-1.
Mbio, 16, 2025
|
|
7CPK
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
5KTX
 
 | | CREBBP bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, CREB-binding protein, ... | | Authors: | Murray, J.M, Noland, C. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
7C62
 
 | |
6R2T
 
 | | Aspergillus niger ferulic acid decarboxylase (Fdc) in complex with prFMN (purified in the radical form) and phenylpropiolic acid | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D. | | Deposit date: | 2019-03-18 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Atomic description of an enzyme reaction dependent on reversible 1,3-dipolar cycloaddition
to be published
|
|
9MZX
 
 | | Crystal structure of human RIPK1 with Compound 1 | | Descriptor: | 1-[(2S,5S)-2,3-dihydro-2,5-methano-1,4-benzoxazepin-4(5H)-yl]-2,2-dimethylpropan-1-one, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Lesburg, C.A, Palte, R.L, Maskos, K, Thomsen, M, Lammens, A. | | Deposit date: | 2025-01-23 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The Discovery of Bridged Benzoazepine Amides as Selective Allosteric Modulators of RIPK1.
Acs Med.Chem.Lett., 16, 2025
|
|
7CPL
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
5MCQ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE AND EXOSITE BINDING PEPTIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, BACE-1 ACTIVE AND EXOSITE BINDING INHIBITOR, Beta-secretase 1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
6R4F
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
