8GMN
 
 | | Crystal structure of human C1s in complex with inhibitor | | Descriptor: | Complement C1s subcomponent, SULFATE ION, [4-(1-aminophthalazin-6-yl)piperazin-1-yl](2-methylphenyl)methanone | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2023-03-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Novel Series of Potent, Selective, Orally Available, and Brain-Penetrable C1s Inhibitors for Modulation of the Complement Pathway.
J.Med.Chem., 66, 2023
|
|
8PSX
 
 | |
8PSZ
 
 | |
6WZM
 
 | | LY3041658 Fab bound to CXCL8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-8, LY3041658 Fab heavy chain, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
8IWD
 
 | | Aspergillus niger Rha-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.J, Li, L, Wang, M.H, Jiang, Z.D, Zhu, Y.B, Jin, T.C, Ni, H. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure and catalytic function of alpha-L-rhamnosidase from Aspergillus niger
To Be Published
|
|
8DLA
 
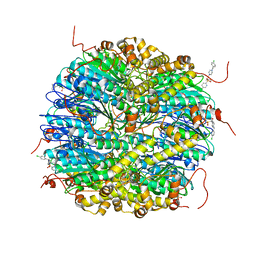 | | ClpP2 from Chlamydia trachomatis bound by MAS1-12 | | Descriptor: | 1-{4-[(4-chlorophenyl)methyl]piperazin-1-yl}-2-methyl-2-[5-(trifluoromethyl)pyridine-2-sulfonyl]propan-1-one, ATP-dependent Clp protease proteolytic subunit 2, GLYCEROL, ... | | Authors: | Azadmanesh, J, Struble, L.R, Seleem, M.A, Ouellette, S, Conda-Sheridan, M, Borgstahl, G.E.O. | | Deposit date: | 2022-07-07 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The structure of caseinolytic protease subunit ClpP2 reveals a functional model of the caseinolytic protease system from Chlamydia trachomatis.
J.Biol.Chem., 299, 2023
|
|
7MKJ
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to T7A1 promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKD
 
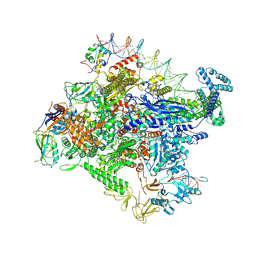 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 1) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OO0
 
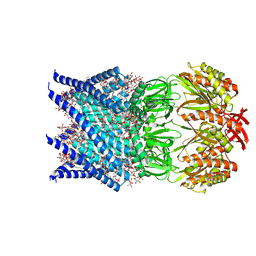 | | Mechanosensitive channel MscS solubilized with DDM in open conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel MscS | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WKK
 
 | | Phage G gp27 major capsid proteins and gp26 decoration proteins | | Descriptor: | Gp26 capsid decoration protein, Gp27 major capsid protein | | Authors: | Monroe, L, Gonzalez, B, Jiang, W, Kihara, D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Phage G Structure at 6.1 angstrom Resolution, Condensed DNA, and Host Identity Revision to a Lysinibacillus.
J.Mol.Biol., 432, 2020
|
|
6O34
 
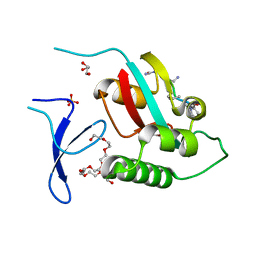 | | Crystal Structure Analysis of PIN1 | | Descriptor: | GLYCEROL, NONAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure Analysis of PIN1
To be Published
|
|
7M3Z
 
 | | Structure of TIM-3 in complex with N-(4-(8-chloro-2-mehtyl-5-oxo-5,6-dihydro-[1,2,4]triazolo[1,5-c]quinazolin-9-yl)-3-methylphenyl)methanesulfonamdide (compound 35) | | Descriptor: | CALCIUM ION, Hepatitis A virus cellular receptor 2, N-{4-[(4S,10aP)-8-chloro-2-methyl-5-oxo-5,6-dihydro[1,2,4]triazolo[1,5-c]quinazolin-9-yl]-3-methylphenyl}methanesulfonamide | | Authors: | Rietz, T.A. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment-Based Discovery of Small Molecules Bound to T-Cell Immunoglobulin and Mucin Domain-Containing Molecule 3 (TIM-3).
J.Med.Chem., 64, 2021
|
|
7MIF
 
 | | Human CTPS1 bound to inhibitor R80 | | Descriptor: | CTP synthase 1, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIP
 
 | | Mouse CTPS1 bound to inhibitor R80 | | Descriptor: | CTP synthase 1, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YHG
 
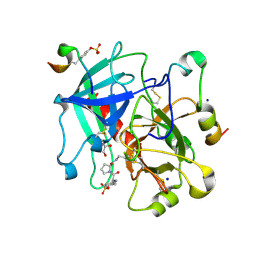 | | Thrombin in complex with D-Phe-Pro-m-methoxybenzylamide derivative (16a) | | Descriptor: | 1-(3-methoxyphenyl)methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, D-PHENYLALANINE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G, Collins, C. | | Deposit date: | 2020-03-29 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Thrombin in complex with D-Phe-Pro-m-methoxybenzylamide derivative (16a)
To be published
|
|
6WUW
 
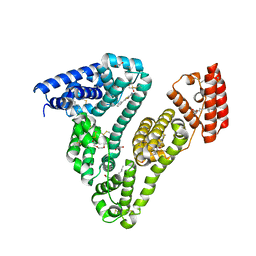 | | Crystal structure of Human Serum Albumin complex with JMS-053 | | Descriptor: | 1,2-ETHANEDIOL, 7-imino-2-phenylthieno[3,2-c]pyridine-4,6(5H,7H)-dione, MYRISTIC ACID, ... | | Authors: | Czub, M.P, Cooper, D.R, Shabalin, I.G, Lazo, J.S, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Complex of an Iminopyridinedione Protein Tyrosine Phosphatase 4A3 Phosphatase Inhibitor with Human Serum Albumin.
Mol.Pharmacol., 98, 2020
|
|
7OZ1
 
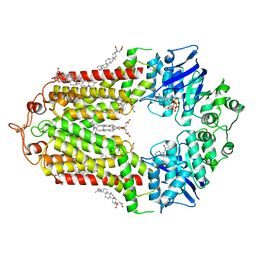 | | Cryo-EM structure of ABCG1 E242Q mutant with ATP and cholesteryl hemisuccinate bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Skarda, L, Kowal, J, Locher, K.P. | | Deposit date: | 2021-06-25 | | Release date: | 2021-09-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Human Cholesterol Transporter ABCG1.
J.Mol.Biol., 433, 2021
|
|
6WM7
 
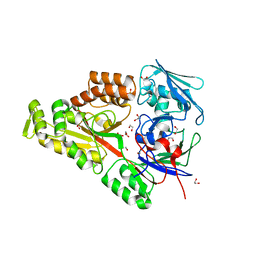 | | Periplasmic EDTA-binding protein EppA, orthorhombic | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein, family 5, ... | | Authors: | Lewis, K.M, Greene, C.L, Sattler, S.A, Xun, L, Kang, C. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The Structural Basis of the Binding of Various Aminopolycarboxylates by the Periplasmic EDTA-Binding Protein EppA from Chelativorans sp. BNC1.
Int J Mol Sci, 21, 2020
|
|
7M7E
 
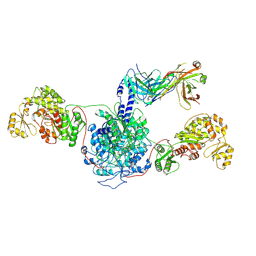 | | 6-Deoxyerythronolide B synthase (DEBS) hybrid module (M3/1) in complex with antibody fragment 1B2 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), 6-deoxyerythronolide-B synthase EryA2, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | Descriptor: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
8GRN
 
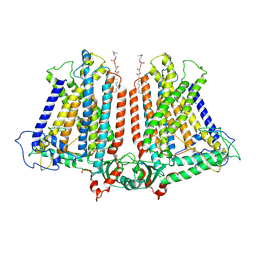 | | AtOSCA1.1 extended state | | Descriptor: | Protein OSCA1, [1-MYRISTOYL-GLYCEROL-3-YL]PHOSPHONYLCHOLINE | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
9BG1
 
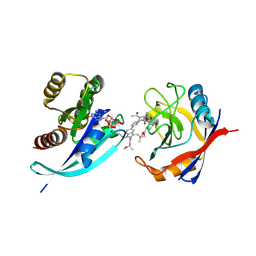 | | Tri-complex of Compound-3, KRAS G12V, and CypA | | Descriptor: | (2R)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1S)-1-methoxyethyl]pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-methyl-2-(N-methylacetamido)butanamide (non-preferred name), GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2024-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Discovery of Daraxonrasib (RMC-6236), a Potent and Orally Bioavailable RAS(ON) Multi-selective, Noncovalent Tri-complex Inhibitor for the Treatment of Patients with Multiple RAS-Addicted Cancers.
J.Med.Chem., 68, 2025
|
|
7MXN
 
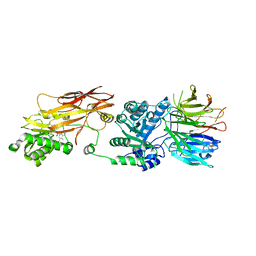 | | PRMT5(M420T mutant):MEP50 complexed with inhibitor PF-06939999 | | Descriptor: | (1S,2S,3S,5R)-3-{[6-(difluoromethyl)-5-fluoro-1,2,3,4-tetrahydroisoquinolin-8-yl]oxy}-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclopentane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7M7H
 
 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1' | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
7M7F
 
 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
