9EPN
 
 | | Crystal structure of HprS histidine kinase cytoplasmic fragment from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, GLYCOCHOLIC ACID, IMIDAZOLE, ... | | Authors: | Koczurowska, A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | Deposit date: | 2024-03-19 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biophysical characterization of the cytoplasmic domains of HprS kinase and its interactions with the cognate regulator HprR.
Arch.Biochem.Biophys., 764, 2024
|
|
4XTJ
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl plus 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5ZXM
 
 | | Crystal Structure of GyraseB N-terminal at 1.93A Resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA gyrase subunit B, ... | | Authors: | Tiwari, P, Gupta, D, Sachdeva, E, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Structural insights into the transient closed conformation and pH dependent ATPase activity of S.Typhi GyraseB N- terminal domain.
Arch.Biochem.Biophys., 701, 2021
|
|
6AZR
 
 | | Crystal structure of the T264A HK853cp-BeF3-RR468 complex | | Descriptor: | Chemotaxis regulator-transmits chemoreceptor signals to flagelllar motor components CheY, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-09-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
4WUD
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from no salt condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6ENH
 
 | |
6DK8
 
 | | RetS kinase region without cobalt | | Descriptor: | NICKEL (II) ION, RetS (Regulator of Exopolysaccharide and Type III Secretion) | | Authors: | Mancl, J.M, Schubot, F.D. | | Deposit date: | 2018-05-29 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Helix Cracking Regulates the Critical Interaction between RetS and GacS in Pseudomonas aeruginosa.
Structure, 27, 2019
|
|
6DK7
 
 | | RetS histidine kinase region with cobalt | | Descriptor: | COBALT (II) ION, RetS (Regulator of Exopolysaccharide and Type III Secretion) | | Authors: | Mancl, J.M, Schubot, F.D. | | Deposit date: | 2018-05-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Helix Cracking Regulates the Critical Interaction between RetS and GacS in Pseudomonas aeruginosa.
Structure, 27, 2019
|
|
6ENG
 
 | | Crystal structure of the 43K ATPase domain of Escherichia coli gyrase B in complex with an aminocoumarin | | Descriptor: | CHLORIDE ION, Coumermycin A1, DNA gyrase subunit B, ... | | Authors: | Vanden Broeck, A, McEwen, A.G, Lamour, V. | | Deposit date: | 2017-10-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Gyrase Interaction with Coumermycin A1.
J.Med.Chem., 62, 2019
|
|
3A0R
 
 | | Crystal structure of histidine kinase ThkA (TM1359) in complex with response regulator protein TrrA (TM1360) | | Descriptor: | MERCURY (II) ION, Response regulator, Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
7YQ8
 
 | | Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, 50-mer DNA, DNA topoisomerase 2-beta, ... | | Authors: | Naganuma, M, Ehara, H, Kim, D, Nakagawa, R, Cong, A, Bu, H, Jeong, J, Jang, J, Schellenberg, M.J, Bunch, H, Sekine, S. | | Deposit date: | 2022-08-05 | | Release date: | 2024-01-03 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ERK2-topoisomerase II regulatory axis is important for gene activation in immediate early genes.
Nat Commun, 14, 2023
|
|
7ZBG
 
 | | Human Topoisomerase II Beta ATPase ADP | | Descriptor: | 4-[(2S,3R)-3-[3,5-bis(oxidanylidene)piperazin-1-ium-1-yl]butan-2-yl]piperazin-4-ium-2,6-dione, ADENOSINE-5'-DIPHOSPHATE, DNA topoisomerase 2-beta, ... | | Authors: | Ling, E.M, Basle, A, Cowell, I.G, Blower, T.R, Austin, C.A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A comprehensive structural analysis of the ATPase domain of human DNA topoisomerase II beta bound to AMPPNP, ADP, and the bisdioxopiperazine, ICRF193.
Structure, 30, 2022
|
|
7ZP0
 
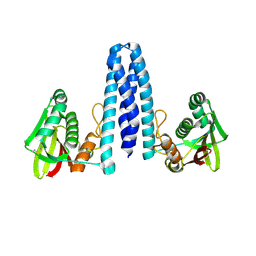 | |
7Z8N
 
 | | GacS histidine kinase from Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, Histidine kinase, R-1,2-PROPANEDIOL | | Authors: | Fadel, F, Bassim, V, Francis, V.I, Porter, S.L, Botzanowski, T, Legrand, P, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-03-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
7Z9C
 
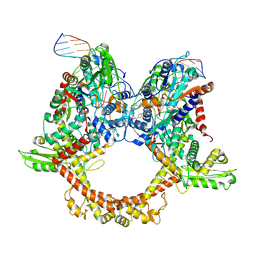 | | E.coli gyrase holocomplex with 217 bp DNA and albicidin | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*AP*CP*CP*CP*GP*CP*AP*CP*AP*GP*AP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*TP*TP*AP*TP*GP*CP*CP*TP*GP*AP*TP*TP*CP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*CP*TP*GP*TP*GP*CP*GP*GP*GP*T)-3'), ... | | Authors: | Ghilarov, D, Heddle, J.G.H, Suessmuth, R. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin.
Nat Catal, 6, 2023
|
|
7Z9M
 
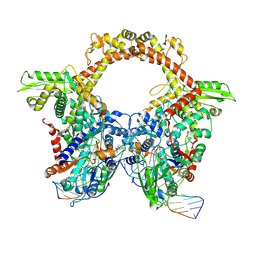 | | E.coli gyrase holocomplex with 217 bp DNA and Albi-1 (site AA) | | Descriptor: | 4-[[4-[[5-[[(2S)-2-[[5-[(4-cyanophenyl)carbonylamino]pyridin-2-yl]carbonylamino]-3-(1H-1,2,3-triazol-4-yl)propanoyl]amino]pyridin-2-yl]carbonylamino]-2-oxidanyl-3-propan-2-yloxy-phenyl]carbonylamino]benzoic acid, DNA (5'-D(*AP*AP*TP*CP*AP*CP*CP*CP*GP*CP*AP*CP*AP*GP*AP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*TP*TP*AP*TP*GP*CP*CP*TP*GP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Ghilarov, D, Heddle, J.G.H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin.
Nat Catal, 6, 2023
|
|
7Z9G
 
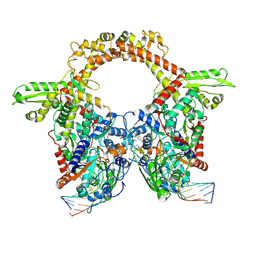 | | E.coli gyrase holocomplex with 217 bp DNA and Albi-2 | | Descriptor: | 4-[[3-(2-azanylethoxy)-2-oxidanyl-4-[[5-[[(2~{S})-2-[[4-[(6-oxidanylnaphthalen-2-yl)carbonylamino]phenyl]carbonylamino]-3-(1~{H}-1,2,3-triazol-4-yl)propanoyl]amino]pyridin-2-yl]carbonylamino]phenyl]carbonylamino]-3-methoxy-2-oxidanyl-benzoic acid, DNA (5'-D(*AP*AP*TP*CP*AP*CP*CP*CP*GP*CP*AP*CP*AP*GP*AP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*TP*TP*AP*TP*GP*CP*CP*TP*GP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Ghilarov, D, Heddle, J.G.H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin.
Nat Catal, 6, 2023
|
|
7Z9K
 
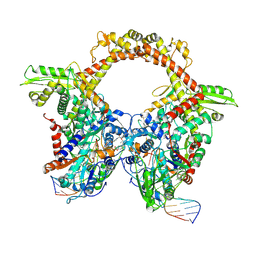 | | E.coli gyrase holocomplex with 217 bp DNA and Albi-1 (site TG) | | Descriptor: | 4-[[4-[[5-[[(2S)-2-[[5-[(4-cyanophenyl)carbonylamino]pyridin-2-yl]carbonylamino]-3-(1H-1,2,3-triazol-4-yl)propanoyl]amino]pyridin-2-yl]carbonylamino]-2-oxidanyl-3-propan-2-yloxy-phenyl]carbonylamino]benzoic acid, DNA (5'-D(*AP*AP*TP*CP*AP*CP*CP*CP*GP*CP*AP*CP*AP*GP*AP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*TP*TP*AP*TP*GP*CP*CP*TP*GP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Ghilarov, D, Heddle, J.G.H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin.
Nat Catal, 6, 2023
|
|
8A52
 
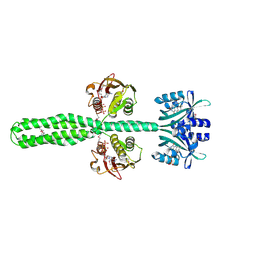 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c-axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Putative Sensory box protein,Sensor protein FixL | | Authors: | Batra-Safferling, R, Arinkin, V, Granzin, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
8A6X
 
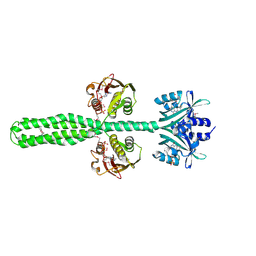 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Sensor protein FixL | | Authors: | Arinkin, V, Granzin, J, Batra-Safferling, R. | | Deposit date: | 2022-06-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
8A7F
 
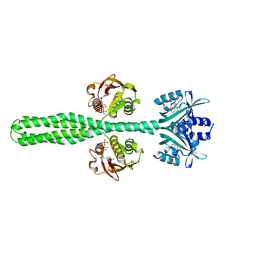 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (asymmetrical variant, trigonal form with long c axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Sensor protein FixL | | Authors: | Batra-Safferling, R, Arinkin, V, Granzin, J. | | Deposit date: | 2022-06-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
8A7H
 
 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (light state; asymmetrical variant, trigonal form with long c axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Flavin mononucleotide (semi-quinone intermediate), Putative Sensory box protein,Sensor protein FixL | | Authors: | Arinkin, V, Granzin, J, Batra-Safferling, R. | | Deposit date: | 2022-06-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.145 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (light state; asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
8A3U
 
 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (symmetrical variant, trigonal form with short c-axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Sensor protein FixL | | Authors: | Arinkin, V, Batra-Safferling, R, Granzin, J. | | Deposit date: | 2022-06-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (symmetrical variant, trigonal form
with short c-axis)
To Be Published
|
|
8AVX
 
 | |
8AVW
 
 | | Cryo-EM structure of DrBphP in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome,Response regulator | | Authors: | Wahlgren, W.Y, Takala, H, Westenhoff, S. | | Deposit date: | 2022-08-27 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural mechanism of signal transduction in a phytochrome histidine kinase.
Nat Commun, 13, 2022
|
|
