1VIJ
 
 | | HIV-1 PROTEASE COMPLEXED WITH THE INHIBITOR HOE/BAY 793 HEXAGONAL FORM | | Descriptor: | HIV-1 PROTEASE, N-(1-BENZYL-2,3-DIHYDROXY-4-{3-METHYL-2-[2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYLAMINO]-BUTYRYLAMINO}-5-PHENYL-PENTYL)-3-METHYL-2-[2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYLAMINO]-BUTYRAMIDE | | Authors: | Lange-Savage, G, Berchtold, H, Liesum, A, Hilgenfeld, R. | | Deposit date: | 1997-05-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of HOE/BAY 793 complexed to human immunodeficiency virus (HIV-1) protease in two different crystal forms--structure/function relationship and influence of crystal packing.
Eur.J.Biochem., 248, 1997
|
|
1VIK
 
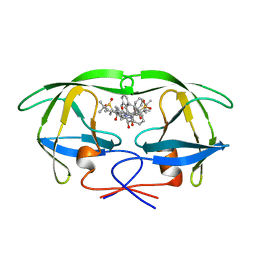 | | HIV-1 PROTEASE COMPLEXED WITH THE INHIBITOR HOE/BAY 793 ORTHORHOMBIC FORM | | Descriptor: | HIV-1 PROTEASE, N-(1-BENZYL-2,3-DIHYDROXY-4-{3-METHYL-2-[2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYLAMINO]-BUTYRYLAMINO}-5-PHENYL-PENTYL)-3-METHYL-2-[2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYLAMINO]-BUTYRAMIDE | | Authors: | Lange-Savage, G, Berchtold, H, Liesum, A, Hilgenfeld, R. | | Deposit date: | 1997-05-07 | | Release date: | 1998-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of HOE/BAY 793 complexed to human immunodeficiency virus (HIV-1) protease in two different crystal forms--structure/function relationship and influence of crystal packing.
Eur.J.Biochem., 248, 1997
|
|
1W5X
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,3-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1NPW
 
 | | Crystal structure of HIV protease complexed with LGZ479 | | Descriptor: | CARBAMIC ACID 1-{5-BENZYL-5-[2-HYDROXY-4-PHENYL-3-(TETRAHYDRO-FURAN- 3-YLOXYCARBONYLAMINO)-BUTYL]-4-OXO-4,5-DIHYDRO-1H-PYRROL-3-YL}- INDAN-2-YL ESTER, POL polyprotein | | Authors: | Smith III, A.B. | | Deposit date: | 2003-01-20 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors.
J.Med.Chem., 46, 2003
|
|
1W5V
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DI-3-FLUORO-BENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1ODX
 
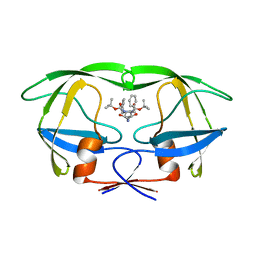 | | HIV-1 Proteinase mutant A71T, V82A | | Descriptor: | HIV-1 PROTEASE, di-tert-butyl {iminobis[(2S,3S)-3-hydroxy-1-phenylbutane-4,2-diyl]}biscarbamate | | Authors: | Kervinen, J, Thanki, N, Zdanov, A, Wlodawer, A. | | Deposit date: | 1996-09-16 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Native and Drug-Resistant HIV-1 Proteinases Complexed with an Aminodiol Inhibitor
Protein Pept.Lett., 3, 1996
|
|
1ODW
 
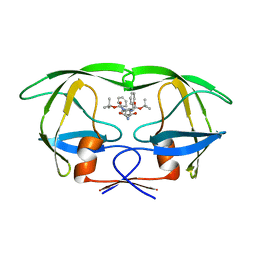 | | Native HIV-1 Proteinase | | Descriptor: | HIV-1 PROTEASE, di-tert-butyl {iminobis[(2S,3S)-3-hydroxy-1-phenylbutane-4,2-diyl]}biscarbamate | | Authors: | Thanki, N, Kervinen, J, Wlodawer, A. | | Deposit date: | 1996-09-16 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of the Native and Drug-Resistant HIV-1 Proteinases Complexed with an Aminodiol Inhibitor
Protein Pept.Lett., 3, 1996
|
|
1OHR
 
 | |
1Z1R
 
 | | HIV-1 protease complexed with Macrocyclic peptidomimetic inhibitor 2 | | Descriptor: | 2-[(8S,11S)-11-{(1R)-1-HYDROXY-2-[ISOPENTYL(PHENYLSULFONYL)AMINO]ETHYL}-6,9-DIOXO-2-OXA-7,10-DIAZABICYCLO[11.2.2]HEPTADECA-1(15),13,16-TRIEN-8-YL]ACETAMIDE, Pol polyprotein, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease
Biochemistry, 38, 1999
|
|
3HLO
 
 | |
3HZC
 
 | |
3JVY
 
 | | HIV-1 Protease Mutant G86A with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3KA2
 
 | |
1NPV
 
 | | Crystal structure of HIV-1 protease complexed with LDC271 | | Descriptor: | POL polyprotein, {1-BENZYL-3-[2-BENZYL-3-OXO-4-(1-OXO-1,2,3,4-TETRAHYDRO- ISOQUINOLIN-4-YL)-2,3-DIHYDRO-1H-PYRROL-2-YL]-2- HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Smith, A.B. | | Deposit date: | 2003-01-20 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors.
J.Med.Chem., 46, 2003
|
|
1ZLF
 
 | | Crystal structure of a complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor | | Descriptor: | N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-glutaminyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIV-1 protease mutations and inhibitor modifications monitored on a series of complexes. Structural basis for the effect of the A71V mutation on the active site
J.Med.Chem., 49, 2006
|
|
1ZPA
 
 | | HIV Protease with Scripps AB-3 Inhibitor | | Descriptor: | Pol polyprotein, TERT-BUTYL 4-[({[1-((1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL)-1H-1,2,3-TRIAZOL-4-YL]METHYL}AMINO)CARBONYL]BENZYLCARBAMATE | | Authors: | Brik, A, Alexandratos, J, Lin, Y.C, Elder, J.H, Olson, A.J, Wlodawer, A, Goodsell, D.S, Wong, C.H. | | Deposit date: | 2005-05-16 | | Release date: | 2005-05-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors
Chembiochem, 6, 2005
|
|
3HVP
 
 | |
3HAW
 
 | |
3H5B
 
 | | Crystal structure of wild type HIV-1 protease with novel P1'-ligand GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tie, Y, Wang, Y.F, Weber, I.T. | | Deposit date: | 2009-04-21 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Design of HIV-1 protease inhibitors with pyrrolidinones and oxazolidinones as novel P1'-ligands to enhance backbone-binding interactions with protease: synthesis, biological evaluation, and protein-ligand X-ray studies.
J.Med.Chem., 52, 2009
|
|
3HBO
 
 | | Crystal structure of chemically synthesized [D-Ala51/51']HIV-1 protease | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, [D-Ala51/51']HIV-1 protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2009-05-04 | | Release date: | 2010-05-26 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1ZP8
 
 | | HIV Protease with inhibitor AB-2 | | Descriptor: | Pol polyprotein, [1-((1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL)-1H-1,2,3-TRIAZOL-4-YL]METHYL (1R,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YLCARBAMATE | | Authors: | Brik, A, Alexandratos, J.N, Elder, J.H, Olson, A.J, Wlodawer, A, Goodsell, D.S, Wong, C.H. | | Deposit date: | 2005-05-16 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors.
Chembiochem, 6, 2005
|
|
1NPA
 
 | | crystal structure of HIV-1 protease-hup | | Descriptor: | (3S)-TETRAHYDROFURAN-3-YL (1R,2S)-3-[4-((1R)-2-{[(S)-AMINO(HYDROXY)METHYL]OXY}-2,3-DIHYDRO-1H-INDEN-1-YL)-2-BENZYL-3-OXOPYRROLIDIN-2-YL]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, POL polyprotein | | Authors: | Smith III, A.B, Hirschmann, R, Pasternak, A, Yao, W, Sprengeler, P.A, Holloway, M.K, Kuo, L.C, Chen, Z, Darke, P.L, Schleif, W.A. | | Deposit date: | 2003-01-17 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable pyrrolinone inhibitor of HIV-1 protease: computational analysis and X-ray crystal structure of the enzyme complex.
J.MED.CHEM., 40, 1997
|
|
3JVW
 
 | | HIV-1 Protease Mutant G86A with symmetric inhibitor DMP323 | | Descriptor: | Gag-Pol polyprotein, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3JW2
 
 | | HIV-1 Protease Mutant G86S with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
1ZPK
 
 | | Crystal structure of the complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[R-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | CHLORIDE ION, N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN, ... | | Authors: | Duskova, J, Skalova, T, Dohnalek, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-17 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mutational Study of Pseudopeptide Inhibitor Binding to HIV-1 Protease; Analysis of Four X-ray Structures
To be Published
|
|
