4REN
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with petunidin | | Descriptor: | 2-(3,4-dihydroxy-5-methoxyphenyl)-3,5,7-trihydroxychromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with delphinidin | | Descriptor: | 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REL
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with kaempferol | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ACETATE ION, GLYCEROL, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
7Y0O
 
 | |
3SDL
 
 | | Crystal structure of human ISG15 in complex with NS1 N-terminal region from influenza B virus, Northeast Structural Genomics Consortium Target IDs HX6481, HR2873, and OR2 | | Descriptor: | Non-structural protein 1, Ubiquitin-like protein ISG15 | | Authors: | Guan, R, Ma, L.-C, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza B virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5JEL
 
 | | Phosphorylated TRIF in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, Phosphorylated TRIF peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEO
 
 | | Phosphorylated Rotavirus NSP1 in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, PHOSPHATE ION, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4P4U
 
 | | Nucleotide-free stalkless-MxA | | Descriptor: | Interferon-induced GTP-binding protein Mx1, SULFATE ION | | Authors: | Rennie, M.L, McKelvie, S.A, Bulloch, E.M, Kingston, R.L. | | Deposit date: | 2014-03-13 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transient Dimerization of Human MxA Promotes GTP Hydrolysis, Resulting in a Mechanical Power Stroke.
Structure, 22, 2014
|
|
4P4S
 
 | | GMPPCP-bound stalkless-MxA | | Descriptor: | Interferon-induced GTP-binding protein Mx1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Rennie, M.L, McKelvie, S.A, Bulloch, E.M, Kingston, R.L. | | Deposit date: | 2014-03-13 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Transient Dimerization of Human MxA Promotes GTP Hydrolysis, Resulting in a Mechanical Power Stroke.
Structure, 22, 2014
|
|
4GL2
 
 | | Structural Basis for dsRNA duplex backbone recognition by MDA5 | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(*AP*UP*CP*CP*GP*CP*GP*GP*CP*CP*CP*U)-3'), ... | | Authors: | Wu, B, Hur, S. | | Deposit date: | 2012-08-13 | | Release date: | 2013-01-09 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (3.557 Å) | | Cite: | Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5.
Cell(Cambridge,Mass.), 152, 2013
|
|
5W5I
 
 | | Human IFIT1 dimer with PPP-AAAA | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1, RNA (5'-D(*(ATP))-R(P*AP*AP*A)-3') | | Authors: | Abbas, Y.M, Martinez-Montero, S, Damha, M.J, Nagar, B. | | Deposit date: | 2017-06-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into IFIT1 dimerization and conformational changes associated with mRNA binding
To Be Published
|
|
5W5H
 
 | | Human IFIT1 dimer with m7Gppp-AAAA | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1, RNA (5'-D(*(GTA))-R(P*AP*AP*A)-3') | | Authors: | Abbas, Y.M, Martinez-Montero, S, Damha, M.J, Nagar, B. | | Deposit date: | 2017-06-15 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into IFIT1 dimerization and conformational changes associated with mRNA binding
To Be Published
|
|
2N00
 
 | | NMR Solution structure of AIM2 PYD from Mus musculus | | Descriptor: | Interferon-inducible protein AIM2 | | Authors: | Hou, X, Niu, X. | | Deposit date: | 2015-03-01 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of AIM2 PYD domain from Mus musculus reveals a distinct alpha 2-alpha 3 helix conformation from its human homologues
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4J0U
 
 | | Crystal structure of IFIT5/ISG58 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5 | | Authors: | Liu, Y, Liang, H, Feng, F, Yuan, L, Wang, Y.E, Crowley, C, Lv, Z, Li, J, Zeng, S, Cheng, G. | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Crystal Structure of IFIT5
To be Published
|
|
3GA3
 
 | | Crystal structure of the C-terminal domain of human MDA5 | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, MDA5, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2009-02-16 | | Release date: | 2009-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the C-terminal domain of human MDA5
To be Published
|
|
4JBJ
 
 | | Structural mimicry for functional antagonism | | Descriptor: | Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Ma, F, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
3UIU
 
 | | Crystal structure of Apo-PKR kinase domain | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Li, F, Li, S, Yang, X, Shen, Y, Zhang, T. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of Apo-PKR kinase domain
TO BE PUBLISHED
|
|
3B6E
 
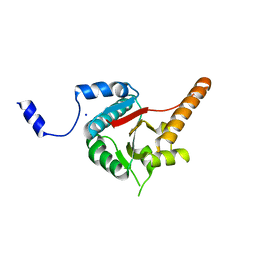 | | Crystal structure of human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SODIUM ION | | Authors: | Karlberg, T, Welin, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain.
To be Published
|
|
4Q2P
 
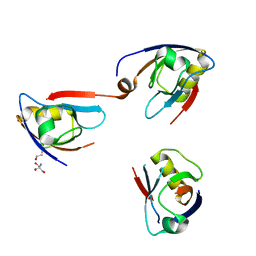 | | NHERF3 PDZ2 in Complex with a Phage-Derived Peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | | Authors: | Appleton, B.A, Wiesmann, C. | | Deposit date: | 2014-04-09 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural portrait of the PDZ domain family.
J.Mol.Biol., 426, 2014
|
|
3TS9
 
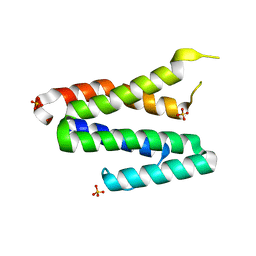 | | Crystal Structure of the MDA5 Helicase Insert Domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SULFATE ION | | Authors: | Berke, I.C, Modis, Y. | | Deposit date: | 2011-09-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA.
Embo J., 31, 2012
|
|
6L4V
 
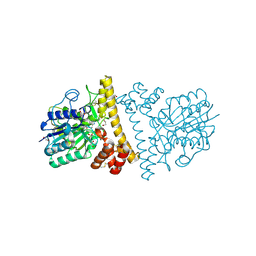 | |
6L4W
 
 | |
6L4Y
 
 | |
6L4X
 
 | |
4OW4
 
 | |
