7X9C
 
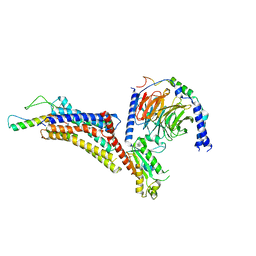 | | Cryo-EM structure of neuropeptide Y Y4 receptor in complex with PP and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Receptor-specific recognition of NPY peptides revealed by structures of NPY receptors.
Sci Adv, 8, 2022
|
|
7X9A
 
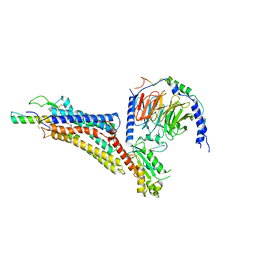 | | Cryo-EM structure of neuropeptide Y Y1 receptor in complex with NPY and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Receptor-specific recognition of NPY peptides revealed by structures of NPY receptors.
Sci Adv, 8, 2022
|
|
7Z6S
 
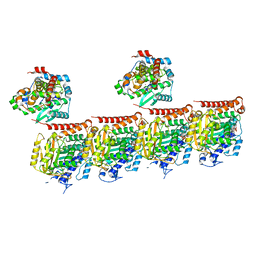 | | MATCAP bound to a human 14 protofilament microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bak, J, Perrakis, A. | | Deposit date: | 2022-03-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z6U
 
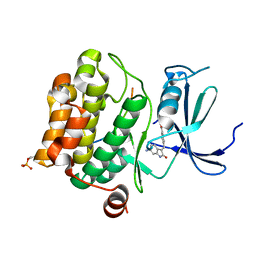 | | Pim1 in complex with (E)-4-((6-amino-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(6-azanyl-2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Isoform 1 of Serine/threonine-protein kinase pim-1, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7UAX
 
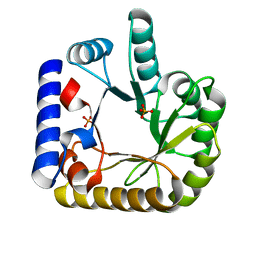 | | The crystal structure of the K36A/K38A double mutant of E. coli YGGS in complex with PLP | | Descriptor: | PHOSPHATE ION, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAU
 
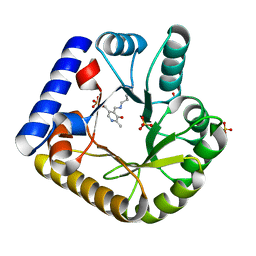 | | The crystal structure of the K137A mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAT
 
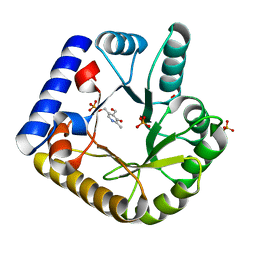 | | The crystal structure of the K36A mutant of E. coli YGGS in complex with PLP | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UB8
 
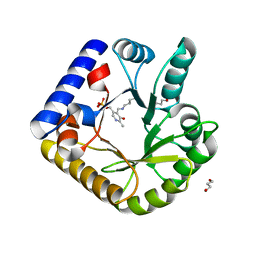 | | The crystal structure of the K38A/K137A/K233A/K234A quadruple mutant of E. coli YGGS in complex with PLP | | Descriptor: | 1,4-BUTANEDIOL, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UB4
 
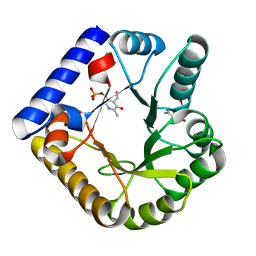 | | The crystal structure of the K36A/K38A/K233A/K234A quadruple mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAZ
 
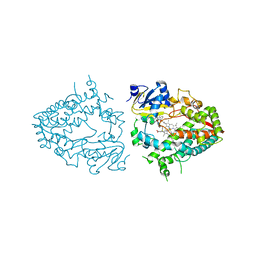 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-{[(pyridin-3-yl)methyl]sulfanyl}propanamide)bis[2-(pyridin-2-yl-kappaN)phenyl-kappaC~1~]iridium, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Ir(III)-Based Agents for Monitoring the Cytochrome P450 3A4 Active Site Occupancy.
Inorg.Chem., 61, 2022
|
|
7UAY
 
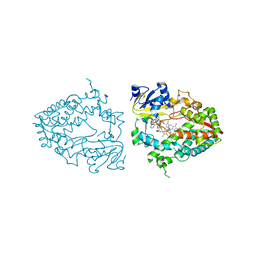 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-{[(pyridin-3-yl)methyl]sulfanyl}propanamide)bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Ir(III)-Based Agents for Monitoring the Cytochrome P450 3A4 Active Site Occupancy.
Inorg.Chem., 61, 2022
|
|
7X8R
 
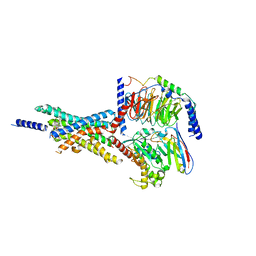 | | Cryo-EM structure of the Boc5-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-[(2-methylpropan-2-yl)oxycarbonylamino]phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X8S
 
 | | Cryo-EM structure of the WB4-24-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-(2-methylpropanoylamino)phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UAI
 
 | | Meprin alpha helix in complex with fetuin-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-13 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
7UAH
 
 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2~{R})-butane-1,2-diol, (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(3-chlorobenzene-1-sulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-03-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
7Z6H
 
 | | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17, Checkpoint protein HUS1, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Nucleic Acids Res., 50, 2022
|
|
7Z6G
 
 | |
7Z6J
 
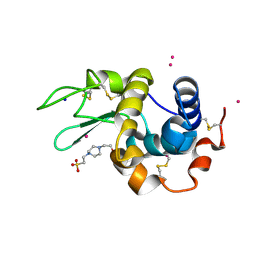 | |
7Z6D
 
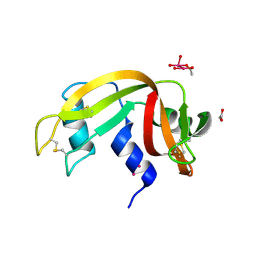 | |
7Z6I
 
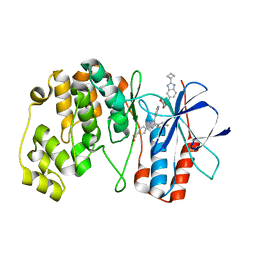 | | Crystal structure of p38alpha C162S in complex with SB20358 and CAS 2094667-81-7 (behind catalytic site; Y35 in), P 21 21 21 | | Descriptor: | 4-[4-(4-fluorophenyl)-2-[4-[methyl(oxidanyl)-$l^{3}-sulfanyl]phenyl]-1~{H}-imidazol-5-yl]pyridine, Mitogen-activated protein kinase 14, N-(2-cyclobutyl-1H-1,3-benzodiazol-5-yl)benzenesulfonamide | | Authors: | Baginski, B, Pous, J, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-03-11 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of p38 alpha autophosphorylation inhibitors that target the non-canonical activation pathway.
Nat Commun, 14, 2023
|
|
7UAA
 
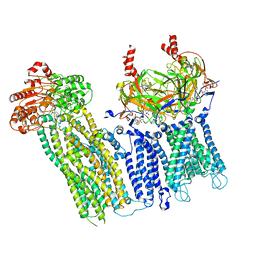 | |
7U9C
 
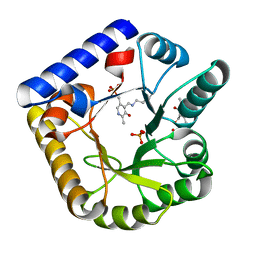 | | Crystal Structure of the wild type Escherichia coli Pyridoxal 5'-phosphate homeostasis protein (YGGS) | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Safo, M.K, Musayev, F.N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7U9H
 
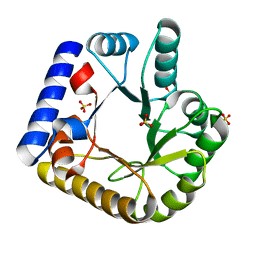 | | Crystal Structure of Escherichia coli apo Pyridoxal 5'-phosphate homeostasis protein (YGGS) | | Descriptor: | Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7X7P
 
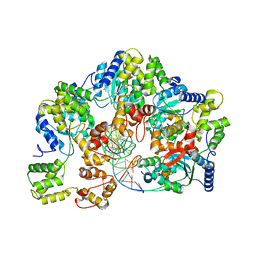 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | Descriptor: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
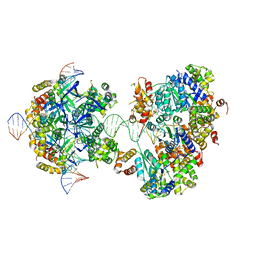 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | Descriptor: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
