8ACC
 
 | | CryoEM structure of sweet potato mild mottle virus VLP | | Descriptor: | Polyprotein, Single-stranded RNA | | Authors: | Javed, A, Byrne, B.M, Ranson, N, Lomonosoff, G. | | Deposit date: | 2022-07-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | CryoEM and stability analysis of virus-like particles of potyvirus and ipomovirus infecting a common host.
Commun Biol, 6, 2023
|
|
6Y9H
 
 | | Thrombin in complex with D-Phe-Pro-m-Trifluoromethylbenzylamide derivative (phe2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-Phe-Pro-m-Trifluoromethylbenzylamide derivative (phe2), DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-03-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Thrombin in complex with D-Phe-Pro-m-Trifluoromethylbenzylamide derivative (phe2)
To be published
|
|
7ZYG
 
 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | Descriptor: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
6YB6
 
 | | Thrombin in complex with D-Phe-Pro-3-chloro-1,3-dihydroxybenzylamide derivative (13c) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-Phe-Pro-3-chloro-1,3-dihydroxybenzylamide derivative, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Thrombin in complex with D-Phe-Pro-3-chloro-1,3-dihydroxybenzylamide derivative (13c)
To be published
|
|
6YHG
 
 | | Thrombin in complex with D-Phe-Pro-m-methoxybenzylamide derivative (16a) | | Descriptor: | 1-(3-methoxyphenyl)methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, D-PHENYLALANINE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G, Collins, C. | | Deposit date: | 2020-03-29 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Thrombin in complex with D-Phe-Pro-m-methoxybenzylamide derivative (16a)
To be published
|
|
6YHJ
 
 | | Thrombin in complex with D-Phe-Pro-2-chlorothiophen derivative (16e) | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(5-chloranylthiophen-2-yl)methyl]pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sandner, A, Heine, A, Klebe, G, Collins, C. | | Deposit date: | 2020-03-30 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Thrombin in complex with D-Phe-Pro-2-chlorothiophen derivative (16e)
To be published
|
|
6YN3
 
 | | Thrombin in complex with 4-hydroxybenzamide (j89) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-HYDROXYBENZAMIDE, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-04-10 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Thrombin in complex with 4-hydroxybenzamide (j89)
To be published
|
|
6YMP
 
 | | Thrombin in complex with 3-((5-(tert-butyl)isoxazol-3-yl)methyl)oxetan-3-amine (j54) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(5-tert-butyl-1,2-oxazol-3-yl)methyl]oxetan-3-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-04-09 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Thrombin in complex with 3-((5-(tert-butyl)isoxazol-3-yl)methyl)oxetan-3-amine (j54)
To be published
|
|
6YSJ
 
 | | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-1-(4-bromophenyl)ethanone, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N, Paulus, A. | | Deposit date: | 2020-04-22 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10)
To be published
|
|
8A8E
 
 | | PPSA C terminal octahedral structure | | Descriptor: | Phosphoenolpyruvate synthase | | Authors: | Song, W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Survival strategies in the heat Lysine acetylation stabilizes the quaternary structure of a Mega-Dalton hyperthermophilic PEP-synthase
To Be Published
|
|
6YTF
 
 | | Acinetobacter baumannii ribosome-tigecycline complex - 30S subunit head | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-24 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
8AKY
 
 | | 290 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKU
 
 | | 250 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKZ
 
 | | 320 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AL0
 
 | | 365 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKX
 
 | | 305 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKV
 
 | | 270 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKW
 
 | | 280 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
6ZH2
 
 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZIT
 
 | | bovine ATP synthase Stator domain, state 2 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C2, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8AXX
 
 | | Expanded Coxsackievirus A9 after treatment with endosomal ionic buffer | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Domanska, A, Plavec, Z, Ruokolainen, V, Loflund, B, Marjomaki, V.S, Butcher, S.J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Studies Reveal that Endosomal Cations Promote Formation of Infectious Coxsackievirus A9 A-Particles, Facilitating RNA and VP4 Release.
J.Virol., 96, 2022
|
|
6ZHX
 
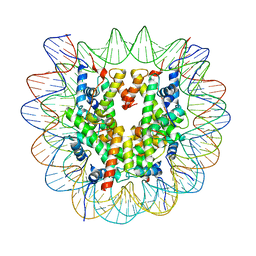 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: nucleosome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (145-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Croll, T.I, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
8AW6
 
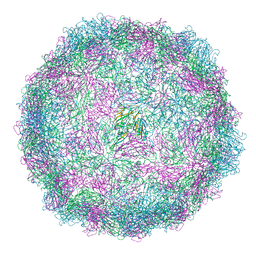 | | Expanded Coxsackievirus A9 after 0.01% faf-BSA treatment | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Domanska, A, Plavec, Z, Ruokolainen, V, Loflund, B, Marjomaki, V.S, Butcher, S.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Studies Reveal that Endosomal Cations Promote Formation of Infectious Coxsackievirus A9 A-Particles, Facilitating RNA and VP4 Release.
J.Virol., 96, 2022
|
|
6YT9
 
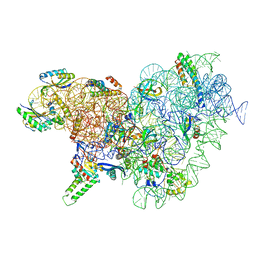 | | Acinetobacter baumannii ribosome-tigecycline complex - 30S subunit body | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-24 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
8AT5
 
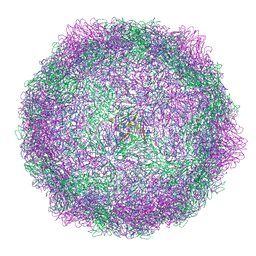 | | native Coxsackievirus A9 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Domanska, A, Plavec, Z, Ruokolainen, V, Marjomaki, V.S, Butcher, S.J. | | Deposit date: | 2022-08-22 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Studies Reveal that Endosomal Cations Promote Formation of Infectious Coxsackievirus A9 A-Particles, Facilitating RNA and VP4 Release.
J.Virol., 96, 2022
|
|
