2AJG
 
 | | Crystal structure of the editing domain of E. coli leucyl-tRNA synthetase | | Descriptor: | Leucyl-tRNA synthetase | | Authors: | Liu, Y, Liao, J, Zhu, B, Wang, E.D, Ding, J. | | Deposit date: | 2005-08-01 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination
Biochem.J., 394, 2006
|
|
2AJH
 
 | | Crystal structure of the editing domain of E. coli leucyl-tRNA synthetase complexes with methionine | | Descriptor: | Leucyl-tRNA synthetase, METHIONINE | | Authors: | Liu, Y, Liao, J, Zhu, B, Wang, E.D, Ding, J. | | Deposit date: | 2005-08-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination
Biochem.J., 394, 2006
|
|
2CYA
 
 | | Crystal structure of tyrosyl-tRNA synthetase from Aeropyrum pernix | | Descriptor: | SULFATE ION, Tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Kuroishi, C, Kuramitsu, S, Terada, T, Bessho, Y, Shirouzu, M, Sekine, S.I, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2005
|
|
2HRA
 
 | | Crystal structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA aminoacylation and nuclear export cofactor Arc1p reveal a novel function for an old fold | | Descriptor: | Glutamyl-tRNA synthetase, cytoplasmic, IODIDE ION | | Authors: | Simader, H, Hothorn, M, Suck, D. | | Deposit date: | 2006-07-20 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA-aminoacylation and nuclear-export cofactor Arc1p reveal a novel function for an old fold.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2CYC
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Pyrococcus horikoshii | | Descriptor: | TYROSINE, tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Sakamoto, K, Terada, T, Shirouzu, M, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
2E3C
 
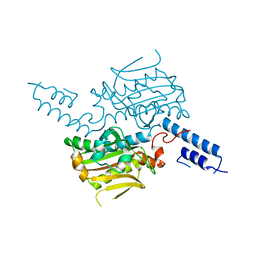 | |
2I4N
 
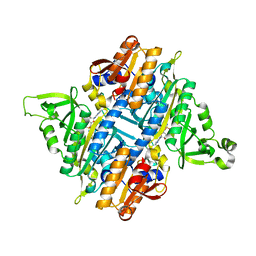 | | Rhodopseudomonas palustris prolyl-tRNA synthetase in complex with CysAMS | | Descriptor: | 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, Proline-tRNA ligase | | Authors: | Crepin, T, Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Two Bacterial Prolyl-tRNA Synthetases with and without a cis-Editing Domain.
Structure, 14, 2006
|
|
2I4M
 
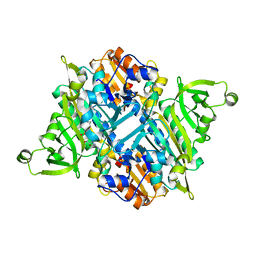 | | Rhodopseudomonas palustris prolyl-tRNA synthetase in complex with ProAMS | | Descriptor: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, Proline-tRNA ligase | | Authors: | Crepin, T, Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Two Bacterial Prolyl-tRNA Synthetases with and without a cis-Editing Domain.
Structure, 14, 2006
|
|
2ZTG
 
 | | Crystal structure of Archaeoglobus fulgidus alanyl-tRNA synthetase lacking the C-terminal dimerization domain in complex with Ala-SA | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, Alanyl-tRNA synthetase, ZINC ION | | Authors: | Naganuma, M, Sekine, S, Fukunaga, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique protein architecture of alanyl-tRNA synthetase for aminoacylation, editing, and dimerization.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7RDN
 
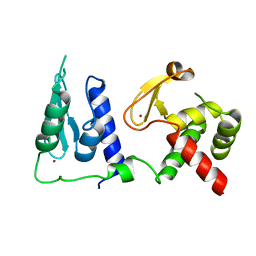 | | Crystal structure of S. cerevisiae pre-mRNA leakage protein 39 (Pml39) | | Descriptor: | Pre-mRNA leakage protein 39, ZINC ION | | Authors: | Hashimoto, H, Ramirez, D.H, Pawlak, N, Blobel, G, Palancade, B, Debler, E.W. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the pre-mRNA leakage 39-kDa protein reveals a single domain of integrated zf-C3HC and Rsm1 modules.
Sci Rep, 12, 2022
|
|
1H4T
 
 | |
2ODR
 
 | | Methanococcus Maripaludis Phosphoseryl-tRNA synthetase | | Descriptor: | phosphoseryl-tRNA synthetase | | Authors: | Steitz, T.A, Kamtekar, S. | | Deposit date: | 2006-12-26 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.228 Å) | | Cite: | Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q5I
 
 | | Crystal structure of apo S581L Glycyl-tRNA synthetase mutant | | Descriptor: | Glycyl-tRNA synthetase, SULFATE ION | | Authors: | Cader, M.Z, Ren, J, James, P.A, Bird, L.E, Talbot, K, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human wildtype and S581L-mutant glycyl-tRNA synthetase, an enzyme underlying distal spinal muscular atrophy.
Febs Lett., 581, 2007
|
|
2PME
 
 | | The Apo crystal Structure of the glycyl-tRNA synthetase | | Descriptor: | Glycyl-tRNA synthetase | | Authors: | Xie, W. | | Deposit date: | 2007-04-21 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Long-range structural effects of a Charcot-Marie- Tooth disease-causing mutation in human glycyl-tRNA synthetase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7AP4
 
 | | Thermus thermophilus Aspartyl-tRNA Synthetase in Complex with Compound AspS7HMDDA | | Descriptor: | (3~{S})-3-azanyl-4-[[(2~{R},3~{S},4~{R},5~{R})-5-[7-azanyl-5-(hydroxymethyl)benzimidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxysulfonylamino]-4-oxidanylidene-butanoic acid, Aspartate--tRNA(Asp) ligase | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Synthesis and Biological Evaluation of 1,3-Dideazapurine-Like 7-Amino-5-Hydroxymethyl-Benzimidazole Ribonucleoside Analogues as Aminoacyl-tRNA Synthetase Inhibitors.
Molecules, 25, 2020
|
|
2Q7H
 
 | | Pyrrolysyl-tRNA synthetase bound to adenylated pyrrolysine and pyrophosphate | | Descriptor: | (2R)-2-AMINO-6-({[(2S,3R)-3-METHYLPYRROLIDIN-2-YL]CARBONYL}AMINO)HEXANOYL [(2S,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL HYDROGEN (R)-PHOSPHATE, 1,2-ETHANEDIOL, PYROPHOSPHATE 2-, ... | | Authors: | Kavran, J.M, Steitz, T.A. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7AP1
 
 | | Klebsiella pneumoniae Seryl-tRNA synthetase in Complex with Compound SerS7HMDDA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Serine-tRNA ligase, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-10-15 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Synthesis and Biological Evaluation of 1,3-Dideazapurine-Like 7-Amino-5-Hydroxymethyl-Benzimidazole Ribonucleoside Analogues as Aminoacyl-tRNA Synthetase Inhibitors.
Molecules, 25, 2020
|
|
2Q5H
 
 | | Crystal structure of apo-wildtype Glycyl-tRNA synthetase | | Descriptor: | Glycyl-tRNA synthetase | | Authors: | Cader, M.Z, Ren, J, James, P.A, Bird, L.E, Talbot, K, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human wildtype and S581L-mutant glycyl-tRNA synthetase, an enzyme underlying distal spinal muscular atrophy.
Febs Lett., 581, 2007
|
|
3O0A
 
 | | Crystal structure of the wild type CP1 hydrolitic domain from Aquifex Aeolicus leucyl-trna | | Descriptor: | Leucyl-tRNA synthetase subunit alpha | | Authors: | Cura, V, Olieric, N, Wang, E.-D, Moras, D, Eriani, G, Cavarelli, J. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the Wild Type and Two Mutants of the Cp1 Hydrolytic Domain from Aquifex Aeolicus Leucyl-tRNA Synthetase
To be Published
|
|
1HTT
 
 | | HISTIDYL-TRNA SYNTHETASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, HISTIDINE, HISTIDYL-TRNA SYNTHETASE | | Authors: | Arnez, J.G, Harris, D.C, Mitschler, A, Rees, B, Francklyn, C.S, Moras, D. | | Deposit date: | 1996-03-09 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate.
EMBO J., 14, 1995
|
|
1GPJ
 
 | | Glutamyl-tRNA Reductase from Methanopyrus kandleri | | Descriptor: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, CITRIC ACID, GLUTAMIC ACID, ... | | Authors: | Moser, J, Schubert, W.-D, Beier, V, Bringemeier, I, Jahn, D, Heinz, D.W. | | Deposit date: | 2001-11-05 | | Release date: | 2002-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis.
EMBO J., 20, 2001
|
|
1WK8
 
 | |
1WKA
 
 | |
1X55
 
 | | Crystal structure of asparaginyl-tRNA synthetase from Pyrococcus horikoshii complexed with asparaginyl-adenylate analogue | | Descriptor: | 5'-O-[N-(L-ASPARAGINYL)SULFAMOYL]ADENOSINE, Asparaginyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Iwasaki, W, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Water-assisted Asparagine Recognition by Asparaginyl-tRNA Synthetase.
J.Mol.Biol., 360, 2006
|
|
1X54
 
 | | Crystal structure of asparaginyl-tRNA synthetase from Pyrococcus horikoshii complexed with asparaginyl-adenylate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-AMINO-1,4-DIOXOBUTAN-2-AMINIUM ADENOSINE-5'-MONOPHOSPHATE, Asparaginyl-tRNA synthetase, ... | | Authors: | Iwasaki, W, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis of the Water-assisted Asparagine Recognition by Asparaginyl-tRNA Synthetase.
J.Mol.Biol., 360, 2006
|
|
