9LE6
 
 | | Composite map of UCC118 Rool RNA | | Descriptor: | UCC118 Rool RNA | | Authors: | Zhang, K, Li, S. | | Deposit date: | 2025-01-07 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structural insights into higher-order natural RNA-only multimers.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9M78
 
 | | Consensus map of UCC118 Rool RNA at 2.26-angstrom resolution | | Descriptor: | UCC118 Rool RNA | | Authors: | Zhang, S, Yi, R, An, L, Li, S, Zhang, K. | | Deposit date: | 2025-03-10 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Structural insights into higher-order natural RNA-only multimers.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9LE5
 
 | | Focused asymmetric unit of UCC118 Rool RNA | | Descriptor: | UCC118 Rool RNA | | Authors: | Zhang, K, Li, S. | | Deposit date: | 2025-01-07 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structural insights into higher-order natural RNA-only multimers.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9PZH
 
 | |
4U55
 
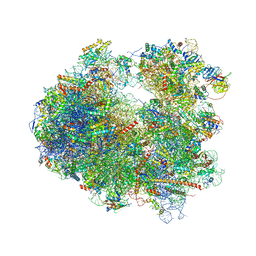 | | Crystal structure of Cryptopleurine bound to the yeast 80S ribosome | | Descriptor: | (14aR)-2,3,6-trimethoxy-11,12,13,14,14a,15-hexahydro-9H-dibenzo[f,h]pyrido[1,2-b]isoquinoline, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
9R59
 
 | | Crystal structure of MAPKAPK2 with a covalent compound GCL334 targeting lysine | | Descriptor: | 5-[3-[bis(oxidanyl)-$l^{3}-sulfanyl]oxy-4-chloranyl-phenyl]-~{N}-(4-piperazin-1-ylphenyl)-~{N}-(pyridin-2-ylmethyl)furan-2-carboxamide, MAP kinase-activated protein kinase 2 | | Authors: | Wang, G.Q, Hillebrand, L, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-05-08 | | Release date: | 2025-07-30 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A twist in the tale: shifting from covalent targeting of a tyrosine in JAK3 to a lysine in MK2.
Rsc Med Chem, 16, 2025
|
|
4U4Z
 
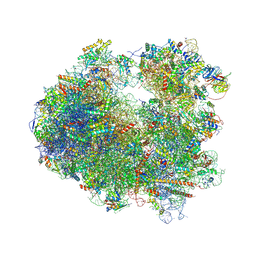 | | Crystal structure of Phyllanthoside bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U4U
 
 | | Crystal structure of Lycorine bound to the yeast 80S ribosome | | Descriptor: | (1S,2S,12bS,12cS)-2,4,5,7,12b,12c-hexahydro-1H-[1,3]dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
8F42
 
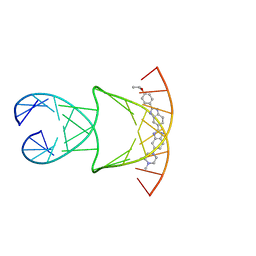 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*TP*TP*AP*AP*GP*GP*AP*AP*TP*TP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8WL5
 
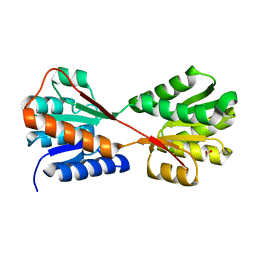 | | X-ray structure of Enterobacter cloacae allose-binding protein in free form | | Descriptor: | 1,2-ETHANEDIOL, Allose ABC transporter | | Authors: | Kamitori, S. | | Deposit date: | 2023-09-29 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray structures of Enterobacter cloacae allose-binding protein in complexes with monosaccharides demonstrate its unique recognition mechanism for high affinity to allose.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
360D
 
 | | STRUCTURE OF 2,5-BIS{[4-(N-ETHYLAMIDINO)PHENYL]}FURAN COMPLEXED TO 5'-D(CPGPCPGPAPAPTPTPCPGPCPG)-3'. A MINOR GROOVE DRUG COMPLEX, SHOWING PATTERNS OF GROOVE HYDRATION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, N,N'-(furan-2,5-diylbis{benzene-4,1-diyl[(Z)-aminomethylylidene]})diethanaminium | | Authors: | Guerri, A, Simpson, I.J, Neidle, S. | | Deposit date: | 1997-10-29 | | Release date: | 1998-07-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Visualisation of extensive water ribbons and networks in a DNA minor-groove drug complex.
Nucleic Acids Res., 26, 1998
|
|
8I43
 
 | |
8I44
 
 | |
8I46
 
 | |
8I45
 
 | |
9V4G
 
 | | Prenyltransferase Ord1 wild type | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, prenyltransferase | | Authors: | Oshiro, T, Uehara, S, Ito, T, Tanaka, Y, Kodera, Y, Matsui, T. | | Deposit date: | 2025-05-23 | | Release date: | 2025-10-01 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Relationship of an All-alpha-helical Prenyltransferase Reveals the Mechanism of Indole Prenylation.
Biochemistry, 64, 2025
|
|
9U53
 
 | |
9OCF
 
 | | 2.73A cryo-EM structure of the Measles Virus L-P in complex with ERdRp-0519 | | Descriptor: | 2-methyl-~{N}-[4-[(2~{S})-2-(2-morpholin-4-ylethyl)piperidin-1-yl]sulfonylphenyl]-5-(trifluoromethyl)pyrazole-3-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Liu, B, Wang, D, Yang, G. | | Deposit date: | 2025-04-24 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis of measles virus polymerase inhibition by nonnucleoside inhibitor ERDRP-0519.
Nat Commun, 16, 2025
|
|
9Q7W
 
 | |
9RM7
 
 | | Crystal Structure of ZIKV NS2B-NS3 protease in complex with ASAP-0036543 | | Descriptor: | (2~{S})-~{N}-[2,2-bis(fluoranyl)ethyl]-2-(2,3-dihydro-1~{H}-isoindol-5-ylamino)-2-[3-methyl-5-(trifluoromethyl)phenyl]ethanamide, DIMETHYL SULFOXIDE, Serine protease subunit NS2B,Serine protease NS3 | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Lee, A, Kenton, N, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Fearon, D, von Delft, F. | | Deposit date: | 2025-06-17 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of ZIKV NS2B-NS3 protease in complex with ASAP-0036543
To Be Published
|
|
9R6O
 
 | |
9OX9
 
 | | Cryo-EM structure of S. Mansoni p97 bound to CB-5083 | | Descriptor: | 1-[4-(benzylamino)-7,8-dihydro-5H-pyrano[4,3-d]pyrimidin-2-yl]-2-methyl-1H-indole-4-carboxamide, vesicle-fusing ATPase | | Authors: | Stephens, D.R, Han, Y, Chen, Z, Collins, J.J, Fung, H.Y.J. | | Deposit date: | 2025-06-03 | | Release date: | 2025-08-13 | | Last modified: | 2025-09-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A genome-scale drug discovery pipeline uncovers therapeutic targets and a unique p97 allosteric binding site in Schistosoma mansoni.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9Y12
 
 | | Crystal Structure of N-Acetyl Transferase Domain-Containing Protein from Bacteroides fragilis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetyltransferase (GNAT) family protein, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2025-08-29 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of N-Acetyl Transferase Domain-Containing Protein from Bacteroides fragilis
To Be Published
|
|
9OCE
 
 | | 2.48A cryo-EM structure of the Measles Virus L-P-C in complex with ERdRp-0519 | | Descriptor: | 2-methyl-~{N}-[4-[(2~{S})-2-(2-morpholin-4-ylethyl)piperidin-1-yl]sulfonylphenyl]-5-(trifluoromethyl)pyrazole-3-carboxamide, Phosphoprotein, Protein C, ... | | Authors: | Liu, B, Wang, D, Yang, G. | | Deposit date: | 2025-04-24 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis of measles virus polymerase inhibition by nonnucleoside inhibitor ERDRP-0519.
Nat Commun, 16, 2025
|
|
9RJR
 
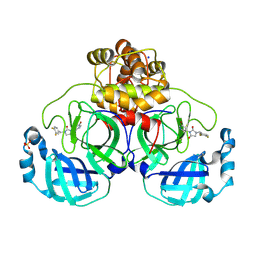 | | SARS-CoV-2 with a bound inhibitor | | Descriptor: | 1-[[3,4-bis(fluoranyl)phenyl]methyl]-3-(5-bromanylpyridin-3-yl)imidazolidine-2,4-dione, 3C-like proteinase nsp5, SODIUM ION, ... | | Authors: | Mac Sweeney, A. | | Deposit date: | 2025-06-12 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | SARS-CoV-2 with a bound inhibitor
To Be Published
|
|
