4UEE
 
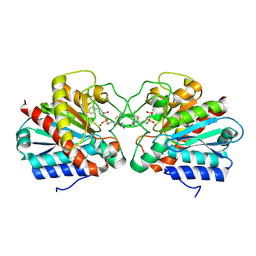 | | Crystal structure of the human CARBOXYPEPTIDASE A1 in complex with the PHOSPHINIC INHIBITOR Acetyl-Leu-Ala-Y(PO2CH2)-homoPhe-OH | | Descriptor: | (2S)-2-{[(R)-{(1R)-1-[(N-acetyl-L-leucyl)amino]ethyl}(hydroxy)phosphoryl]methyl}-4-phenylbutanoic acid, CARBOXYPEPTIDASE A1, ZINC ION | | Authors: | Gallego, P, Covaleda, G, Costenaro, L, Devel, L, Dive, V, Aviles, F.X, Reverter, D. | | Deposit date: | 2014-12-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Human Carboxypeptidase A1 in Complex with Phosphinic Inhibitors
To be Published
|
|
6V70
 
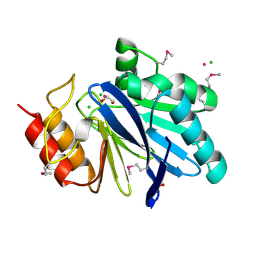 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Cadmium in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CADMIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica.
To Be Published
|
|
3WLJ
 
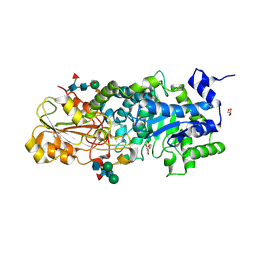 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with 3-deoxy-glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
6UZ3
 
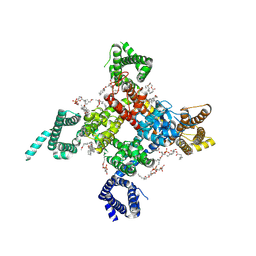 | | Cardiac sodium channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Shi, H, Tonggu, L, Lenaeus, M.J, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Cardiac Sodium Channel.
Cell, 180, 2020
|
|
6I1S
 
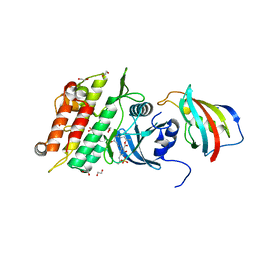 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with FKBP12 and the inhibitor E6201 | | Descriptor: | (4~{S},5~{R},6~{Z},9~{S},10~{S},12~{E})-16-(ethylamino)-4,5-dimethyl-9,10,18-tris(oxidanyl)-3-oxabicyclo[12.4.0]octadeca-1(14),6,12,15,17-pentaene-2,8-dione, 1,2-ETHANEDIOL, Activin receptor type-1, ... | | Authors: | Williams, E.P, Pinkas, D.M, Fortin, J, Newman, J.A, Bradshaw, W.J, Mahajan, P, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-10-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas.
Cancer Cell, 37, 2020
|
|
6V54
 
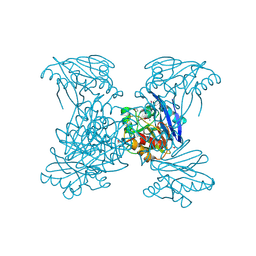 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-03 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica.
To Be Published
|
|
6IIK
 
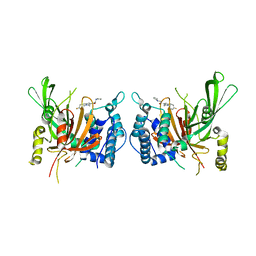 | | USP14 catalytic domain with IU1 | | Descriptor: | 1-[1-(4-fluorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(pyrrolidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, Y.W, He, W, Wang, F. | | Deposit date: | 2018-10-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
7VDR
 
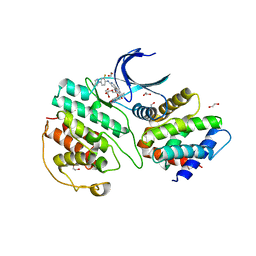 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 13 | | Descriptor: | (1R)-1-[7-[(2-fluoranyl-4-pyrazol-1-yl-phenyl)amino]-1,6-naphthyridin-2-yl]-1-(1-methylpiperidin-4-yl)ethanol, 1,2-ETHANEDIOL, Cyclin-dependent kinase 5 activator 1, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
4UDB
 
 | | MR in complex with desisobutyrylciclesonide | | Descriptor: | DESISOBUYTYRYL CICLESONIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, JellesmarkJensen, T, Cavallin, A, Nilsson, E, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
9E9C
 
 | | Mouse mitoribosome large subunit assembly intermediate bound to NSUN4, METRF4, GTPBP7, GTPBP10 and the MALSU1-L0R8F8-mt-ACP complex (without uL16m), State B1 (SAMC knock-out) | | Descriptor: | 5-cytosine rRNA methyltransferase NSUN4, Acyl carrier protein, mitochondrial, ... | | Authors: | Singh, V, Rorbach, J, Freyer, C, Amunts, A, Wredenberg, A. | | Deposit date: | 2024-11-08 | | Release date: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | The mitochondrial methylation potential gates mitoribosome assembly.
Nat Commun, 16, 2025
|
|
3WEO
 
 | | Sugar beet alpha-glucosidase with acarviosyl-maltohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-09 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
9E8P
 
 | | Crystal Structure of GABARAP-ATG3 conjugate | | Descriptor: | 1,2-ETHANEDIOL, Gamma-aminobutyric acid receptor-associated protein, SULFATE ION, ... | | Authors: | Ohashi, K, Otomo, T. | | Deposit date: | 2024-11-05 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the GABARAP-ATG3 Backside Interaction and Apo ATG3 Conformation.
Biochemistry, 64, 2025
|
|
3BWR
 
 | | SV40 VP1 pentamer in complex with GM1 oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Neu, U, Stehle, T. | | Deposit date: | 2008-01-10 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of GM1 ganglioside recognition by simian virus 40.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6IK5
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
6VCU
 
 | | Homo sapiens FKBP12 protein bound with APX879 in P32 space group | | Descriptor: | ACETATE ION, N'-[(3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,26aS)-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-1,20,21-trioxo-8-(prop-2-en-1-yl)-1,3,4,5,6,8,11,12,13,14,15,16,17,18,19,20,21,23,24,25,26,26a-docosahydro-7H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosin-7-ylidene]acetohydrazide, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Gobeil, S, Spicer, L. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Leveraging Fungal and Human Calcineurin-Inhibitor Structures, Biophysical Data, and Dynamics To Design Selective and Nonimmunosuppressive FK506 Analogs.
Mbio, 12, 2021
|
|
9E9B
 
 | | Crystal structure of L. monocytogenes MenD with Mg2+ and ThDP bound | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Klein, M, Given, F.M, Ho, N.A.T, Allison, T.M, Johnston, J.M. | | Deposit date: | 2024-11-08 | | Release date: | 2025-07-30 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structures of Listeria monocytogenes MenD in ThDP-bound and in-crystallo captured intermediate I-bound forms.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
4ZBV
 
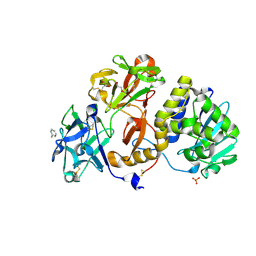 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with benzyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4UE8
 
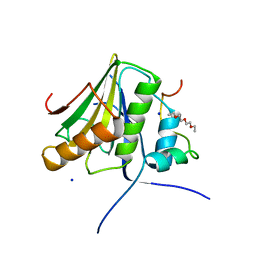 | |
4H1G
 
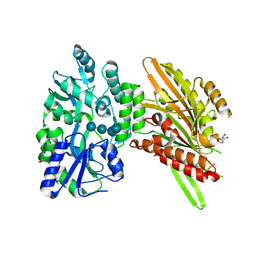 | | Structure of Candida albicans Kar3 motor domain fused to maltose-binding protein | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Allingham, J.A, Duan, D, Delorme, C, Joshi, M. | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the Candida albicans Kar3 kinesin motor domain fused to maltose-binding protein.
Biochem.Biophys.Res.Commun., 428, 2012
|
|
4UDA
 
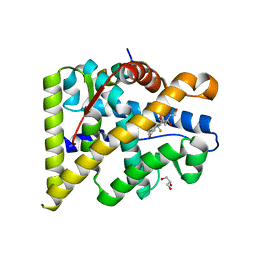 | | MR in complex with dexamethasone | | Descriptor: | DEXAMETHASONE, GLYCEROL, MINERALOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
9EJE
 
 | |
7L8T
 
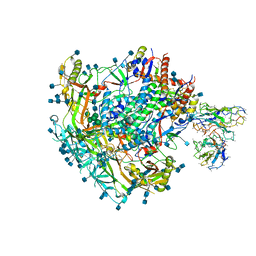 | | BG505 SOSIP.v5.2 N241/N289 in complex with the polyclonal Fab pAbC-1 from animal Rh.33311 (Wk26 time point) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP.v5.2 N241/N289 - gp120, ... | | Authors: | Antanasijevic, A, Sewall, L.M, Ward, A.B. | | Deposit date: | 2020-12-31 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM.
Nat Commun, 12, 2021
|
|
1X8I
 
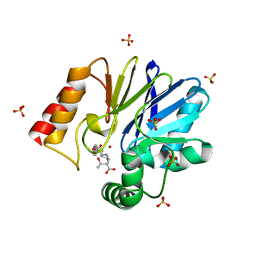 | | Crystal Structure of the Zinc Carbapenemase CphA in Complex with the Antibiotic Biapenem | | Descriptor: | 5H-PYRAZOLO(1,2-A)(1,2,4)TRIAZOL-4-IUM, 6-((2-CARBOXY-6-(1-HYDROXYETHYL)-4-METHYL-7-OXO-1-AZABICYCLO(3.2.0)HEPT-2-EN-3-YL)THIO)-6,7-DIHYDRO-, HYDROXIDE, ... | | Authors: | Garau, G, Dideberg, O. | | Deposit date: | 2004-08-18 | | Release date: | 2004-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Metallo-beta-lactamase Enzyme in Action: Crystal Structures of the Monozinc Carbapenemase CphA and its Complex with Biapenem
J.Mol.Biol., 345, 2005
|
|
9E5M
 
 | | env2 cobalamin riboswitch aptamer domain in complex with ethynyl-4,1'-biphenyl-cobalamin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, N-methylpropane-1,3-diamine, ... | | Authors: | Olenginski, L.T, Batey, R.T. | | Deposit date: | 2024-10-28 | | Release date: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Designing small molecules targeting a cryptic RNA binding site through base displacement.
Nat.Chem.Biol., 2025
|
|
6VEO
 
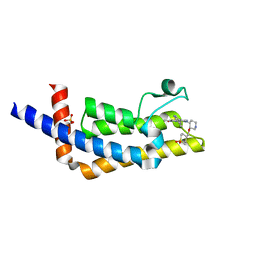 | | ATAD2B bromodomain in complex with 4-({[(3R,4R)-4-{[3-methyl-5-(5-methylpyridin-3-yl)-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl]amino}piperidin-3-yl]oxy}methyl)-1lambda~6~-thiane-1,1-dione (compound 38) | | Descriptor: | 4-({[(3R,4R)-4-{[3-methyl-5-(5-methylpyridin-3-yl)-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl]amino}piperidin-3-yl]oxy}methyl)-1lambda~6~-thiane-1,1-dione, ATPase family AAA domain-containing protein 2B, SULFATE ION | | Authors: | Glass, K.C. | | Deposit date: | 2020-01-02 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Recognition of Mono- and Diacetylated Histones by the ATAD2B Bromodomain.
J.Med.Chem., 63, 2020
|
|
